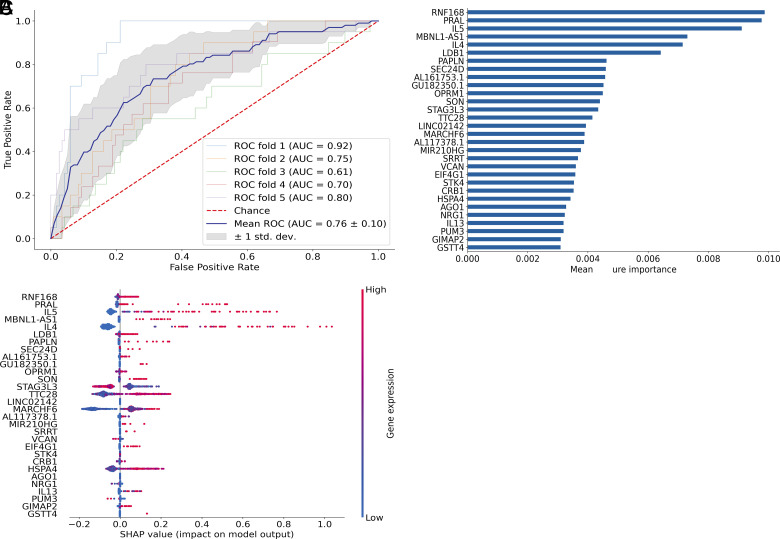
FIGURE 4.
Transcriptional signatures predicted rare env-specific polyfunctional CD4 T cell subsets that had correlated with reduced risk of HIV infection in the RV144 trial. XGBoost machine learning model was used to determine the genes that best predicted the two cytokine profiles associated with reduced risk of infection in Rv144 (correlate profile) versus other env-specific CD4+ T cell subsets (non-correlate profile). (A) Plot shows receiver operating characteristic (ROC) curves created from 5-fold cross-validation. The blue line depicts the mean across the five models. (B) Bar graph of the top 30 most important features based on mean feature importance across the five models. (C) SHAP values showing the contribution of each gene within each cell for the top 30 genes. Color indicates the level of gene expression in each cell for the corresponding gene.