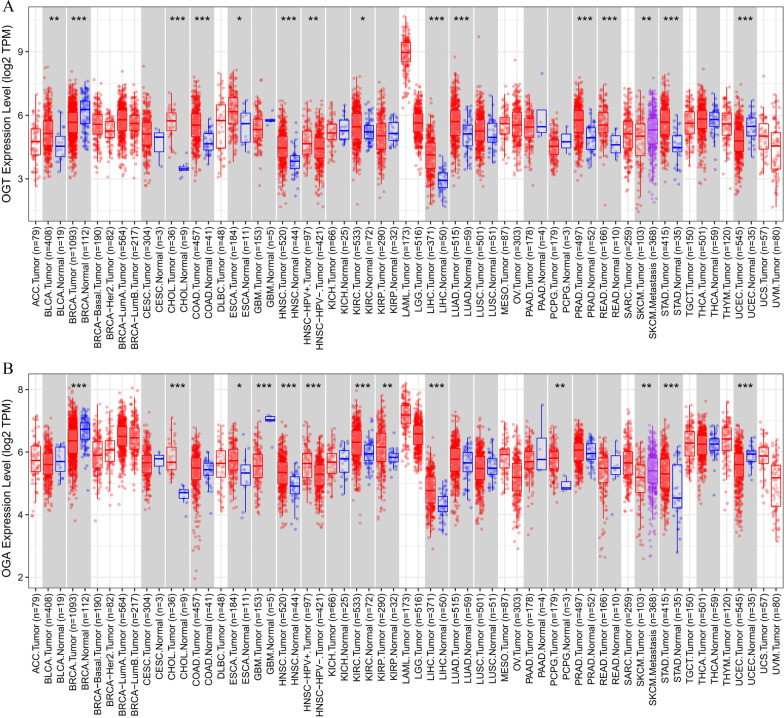
Fig. 3.
Expression profile analyses of O-GlcNAc modifying enzymes across multiple cancers and normal tissues. Expression pattern of OGT (A) and OGA (B) in ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, and UCS. TIMER2.0 was used to generate box plots profiling O-GlcNAc modifying enzymes expression patterns across multiple cancer types (TCGA tumor) and adjacent normal tissue samples (TCGA normal). Each dot represents the individual expression of a distinct tumor or normal sample. The statistical significance computed by the Wilcoxon test is annotated by the number of stars (*p-value < 0.05; **p-value < 0.01; ***p-value < 0.001). As displayed in gray columns when normal data is available