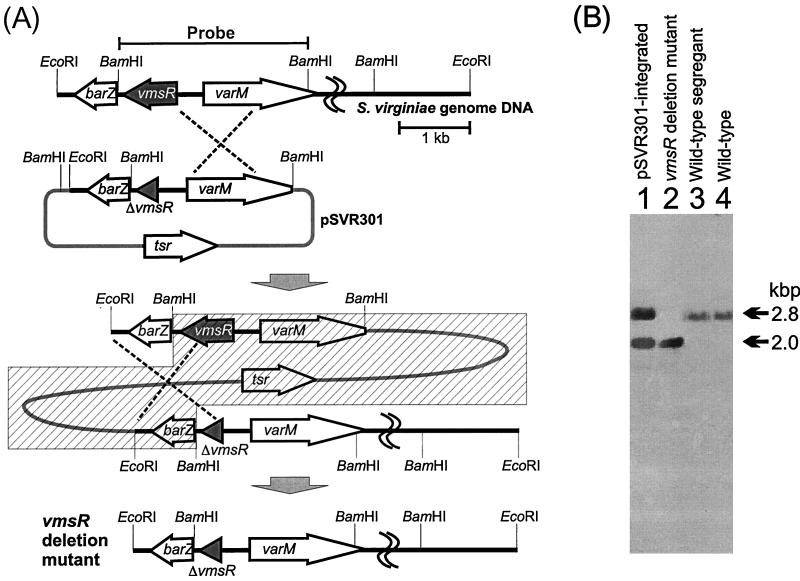
FIG. 1.
Replacement of the S. virginiae vmsR gene with plasmid pSVR301 to form truncated vmsR by homologous recombination. (A) Restriction maps of pSVR301 and the genomic DNA of the S. virginiae wild-type strain. The varM promoter region was amplified by PCR with primers 5′-GGGGCCACTCGCGACTCCAACCGCCGC-3′ (a newly created recognition site for NruI is indicated by underlining) and 5′-GCAAGCGGCTCGCCGTACGGGAGCGCTG-3′ (a BsiWI recognition site is indicated by underlining) using pSVR1 as a template. The NruI-BsiWI-digested PCR fragment was then used to replace the original 1.0-kbp NruI-BsiWI fragment of pSVR1, resulting in the deletion of a 145-bp 5′ untranslated region and the N-terminal 698 bp of vmsR. From the resulting plasmid (pSVR201), a 2.8-kbp EcoRI-HindIII fragment containing barZ, the C-terminal 118 bp of vmsR, and the entire varM gene was cloned into the EcoRI-HindIII site of pUWL-KS to create plasmid pSVR301. A single crossover between pSVR301 and a homologous DNA in the genome gave the pSVR301-integrated strain, and a second crossover generated a vmsR-disrupted strain. Only one of the two possible first crossover events is shown. The hatched box indicates the region deleted by the second crossover. (B) Hybridization patterns of BamHI-digested genomic DNA of the pSVR301-integrated strain (lane 1), vmsR-disrupted strain (lane 2), wild-type segregant (lane 3), and the S. virginiae wild-type strain (lane 4). A 2.8-kbp BamHI fragment containing the vmsR gene was used as a probe. When hybridized with the probe, the vmsR-disrupted strain (lane 2) gave a 2.0-kbp fragment, as a result of the deletion of the vmsR gene.