Abstract
Objective
To evaluate the cytotoxic activity of taraxerol isolated from the leaves of Pterospermum acerifolium and its EtOH extract against human breast, colon, and lung cancer cell lines and docking studies.
Methods
The structures of the isolated compounds were elucidated by several spectroscopic methods such as 1H-NMR, 13C-NMR, DEPT-135, COSY, HSQC, and HMBC. The extract and isolated compound were analyzed for cytotoxic activity on MDA-MB-231, BT-549, A-549, and SW-480 cancer cell lines by MTT assay. Molecular docking was performed on software such as Chem3D pro 12.0.2.1076, Discovery Studio Visualizer, Auto Dock Tools-1.5.6 and Auto dock vina.
Results
The extract and isolated compound taraxerol both displayed excellent inhibitory activity (IC50: 80 µg/mL for extract and 160 µg/mL for compound) on breast cancer cell (MDA-MB-231). The docking studies show a strong affinity with PI3K (-9.8 Kcal/mol) and mTOR (-10.0 Kcal/mol).
Conclusion
The results confirm that the extract and compound exhibited strong cytotoxicity on the MDA-MB-231 cancer cell. So, the extract and the compound may be useful in human chemotherapies.
Keywords: breast cancer, MDA-MB-231, mTOR, PI3K, Pterospermum acerifolium (L.) Willd
1. Introduction
Pterospermum acerifolium (L.) Willd. is commonly known as Kanak Champa, belongs to the family Sterculiaceae and distributed in North Canada, North America, Southeast Asia, Indian sub-Himalayan tract, from India to Burma, and outer Himalayan valley (Kirtikar and Basu, 1933). Traditionally, the use of flowers in the treatment of cancer cell growth (Balachandran & Govindarajan, 2005) and bark in diabetes, smallpox (Caius, 1990), anti-ulcer(Manna, Jena, & Manna, 2009), analgesic (Manna, & Jena, 2009), anti-oxidant (Manna, Manna, & Behera, 2009) and anti-inflammatory activity were reported. Previously, the phytochemical investigation of these species and bark extract showed the presence of kaempferol, luteolin-7-O-glucuronide, kaempferol-3-O-galactoside, kaempferide-7-O-β-D-glucopyranoside, D-galactouronic acid, and acid-polysaccharides, and the flower extract displays β-amyrin and β-sitosterol, etc. Amino acids such as tyrosine, cysteine, glycine, alanine, and leucine were found in trunk bark and seeds (Gupta & Bishnoi, 1979, Gupta et al., 1972, Rizvi and Sultana, 1972). Extract and isolated compound exhibited cytotoxicity on human breast, colon, and lung cancer cell lines in vitro. The compound was investigated theoretically in silico to know targeted pathway.
Phosphoinositide 3-kinases (PI3Ks) are lipid kinases, which plays a regulatory role in cell growth, survival, differentiation, proliferation, and intracellular trafficking (Smith et al., 2012, Vivanco & Sawyers, 2002). It is categorized into three classes (I, II, and III) based on types of regulatory subunits, sequence homology and substrate specificity (Engelman, Luo, & Cantley, 2006). Class I is subdivided into class IA (α, β, δ) and class IB (γ) based on regulatory and catalytic subunit. Class I PI3K family, including four isoforms (P110α, P110β, P110γ and P110δ), is responsible for phosphorylation of phosphatidylinositol (4,5) diphosphate (PIP2) at the 3-position of the inositol ring into phosphatidylinositol (3,4,5) triphosphate (PIP3), which work in the downstream activation for cellular processes as the second messenger (Cantley, 2002). The dephosphorylation of PIP3 into PIP2 by PTEN (phosphatase and tensin homologue gene) serves as a negative regulator and this process is controlled by PTEN (Liu, Cheng, Roberts, & Zhao, 2009). PIP3regulates serine-threonine kinase, AKT, which triggers a cascade singling that ultimately stimulates of mammalian target of rapamycin (mTOR, containing two complexes, mTORC1, and mTORC2). mTOR is a protein kineses of 288.892 kDa serine/threonine that has approximately 2550 amino acid which is encoded by 7650 nucleotides. The mTOR plays two functionally important role as mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Murugan, 2019). A mTORC1 complex consists of PRAS40 (proline-rich AKT substrate 40 kDa), RAPTOR (regulatory associated protein of mTOR) and mLST8 (mammalian lethal with sec13 protein 8), RAPTOR serves as a positive regulator and PRAS40 as a negative regulator for mTOR activity (Haar, Lee, Bandhakavi, Griffin, & Kim, 2007, Hara et al., 2002, Sancak et al., 2007). A mTORC2 consists of mLST8, mSIN1 (mammalian stress-activated protein kinase interacting protein) and RICTOR (rapamycin-insensitive companion of mTOR) (Sarbassov et al., 2004). mTORC1 in downstream phosphorylates and activates of two regulators P70 ribosomal S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) (Proud, 2009). There are two key regulators of ribosome biogenesis and mRNA translation, which plays a vital role in protein synthesis and translation initiation (Manning, 2004). Various studies confirm that external factors such as nutrient intake, growth factors regulate of AKT, mTOR, thereby bypassing PI3K and thus cancer genesis is restarting (Peng, Golub, & Sabatini, 2002, Shaw and Cantley, 2006).Therefore, it is necessary for dual kinase inhibitor compound, which can inhibit PI3K/AKT/mTOR singling pathways. Abnormalities in this signaling pathway are common compared to other major signaling pathways in human cancers (Ediriweera, Tennekoon, & Samarakoon, 2019). Targets of PI3K/AKT/mTOR pathways in the treatment of cancer may be a great strategy. Breast cancer is increasing day by day in the breast tissue of women. A triple-negative breast cancer cell MDA-MB-231 is commonly used in medical laboratories.
This article describes the cytotoxic activity of the extract and isolated compound against human breast (MDA-MB-231 and BT-549), colon (SW-480) and lung (A-549) cancer cell lines and docking studies for inhibitory signaling pathway.
2. Materials and methods
2.1. General information
IR spectral analysis with KBr disc was performed on a Perkin-Elmer 10.4.00 instrument. The Bruker Avance II spectrometer was used to obtain 1H NMR (400 MHz), 13C NMR (100 MHz), HSQC, HMBC, COSY spectra (Bruker BioSpin, Fallanden, Switzerland) in CDCl3 withTMS (tetramethylsilane) as an internal standard. ESI-MS was obtained on XEVO G2-XS QTOF instrument (Water India Pvt Ltd, Bangalore, India). Melting points were measured by Stuart digital melting point apparatus (SPM10) which is uncorrected. All TLC analysis was performed at the precoated TLC glass plate using analytical silica gel GF254 and visualized under UV light (254 and 366 nm) and iodine chamber. Penicillin, Streptomycin, RPMI-1640 medium, 3-(4,5, -dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma –Aldrich (Bangalore, India). FBS (Fetal bovine serum) was acquired from the GIBCO Invitrogen Corporation, USA. All solvents such as benzene, petroleum ether (60–80 °C), chloroform, ethyl acetate, ethanol, dimethylsulfoxide (DMSO) were obtained from E-Merck, India.
2.2. Plant materials
The leaves of P. acerifolium plant were collected from AMU University area Aligarh, UP, India, and sample was identified by plant taxonomy professor M. Badruzzaman Siddiqui, Department of Botany, AMU, Aligarh where voucher specimen No. 31,421 was deposited.
2.3. Extraction and isolation
The shade air-dried and pulverized leaves (2.5 kg) of P. acerifolium plant were exhaustively extracted with 82% EtOH at room temperature for 15 d. The EtOH extract was filtered using a Buchner funnel and concentrated under reduced pressure at 60 °C using rotary evaporators to give a black solid residue (95 g). The dry EtOH crude extract was partitioned by C6H6 and EtOAc solvent to yield benzene crude extract (28 g) and ethyl acetate crude extract (24 g). The C6H6 crude extract (28 g) was subjected initially to silica gel (60–120 mesh) glass column chromatography using petroleum ether to benzene gradient (100:0/0:100, volume percentage) to give 12 main fractions F1 − F12. Each fraction was collected and monitored by TLC. Fraction F8 was chromatographed on a silica gel column, eluted with petroleum ether-benzene (50:50/10:90, volume percentage) to yield four subfractions (F81 − F84). Subfraction F83 was purified by column chromatography, using gradient petroleum ether/C6H6 (30/70) to yield one pure compound as(3β)-D-friedoolean-14-en-3ol (75 mg). The purified compound was characterized by spectral data 1H NMR, 13C NMR, DEPT-135, COSY, HMBC, and HSQC.
2.4. (3β)-D-friedoolean-14-en-3ol (taraxerol)
White crystal (CHCl3); mp 278 °C; ESI-MS (m/z): 427 [M + H]+; IR (KBr) max 3484, 2935.9, 2862.6, 1643.1, 1467.2, 1382.2, 1036.3 cm−1. 1HNMR (CDCl3 , 400 MHZ) δH 0.80 (3H, s, H-24), 0.82 (3H, s, H-26), 0.90 (3H, s, H-28), 0.90 (3H, s, H-30), 0.92 (3H, s, H-25), 0.95 (3H, s, H-29), 0.97 (3H, s, H-23), 1.09 (3H, s, H-27), 3.19 (1H , dd, J = 11.5, 4.64, H-3), 5.53 (1H, dd, J = 3.24, 8.4, H-15). 13C NMR δC 37.71 (C-1), 27.13 (C-2), 79.19 (C-3), 38.97 (C-4), 55.96 (C-5), 18.79 (C-6), 41.33 (C-7), 37.99 (C-8), 48.73 (C-9), 38.76 (C-10), 17.50 (C-11), 35.18 (C-12), 37.56 (C-13), 158.08 (C-14), 116.88 (C-15), 37.73 (C-16), 35.79 (C-17), 49.27 (C-18), 36.68 (C-19), 28.80 (C-20), 33.69 (C-21), 33.09 (C-22), 28.0 (C-23), 15.43 (C-24), 15.46 (C-25), 29.83 (C-26), 25.91 (C-27), 29.93 (C-28), 33.35 (C-29), 21.32 (C-30). HSQC C-1 (37.71, 1.35, 0.98; CH2), C-2 (27.13, 1.58, 1.13; CH2), C-3 (79.19, 3.19; CH), C-5 (55.96, 0.78; CH), C-6 (18.79, 1.73, 1.58; CH2), C-7 (41.33, 2.03, 1.34; CH2), C-9 (48.73, 0.95; CH), C-11 (17.50, 1.65, 1.55; CH2), C-12 (35.18, 1.37, 1.02; CH2), C-15 (116.88, 5.53; CH), C-16 (37.73, 1.91, 1.62; CH2), C-18 (49.27, 1.42; CH), C-19 (36.68, 1.31, 1.02), C-21 (33.69, 1.35, 0.87; CH2), C-22 (33.09, 1.58, 1.26; CH2), C-23 (28.0, 0.98; CH3), C-24 (15.43, 0.80; CH3), C-25 (15.46, 0.92; CH3), C-26 (29.83, 0.82; CH3), C-27 (25.91, 1.09; CH3), C-28 (29.93, 0.90; CH3), C-29 (33.35, 0.95; CH3), C-30 (21.32, 0.90; CH3).
2.5. Cell culture, growth conditions and treatment
Human breast cancer cell lines (MDA-MB-231 and BT-549), a colon cancer cell (SW-480) and lung cell line (A-549) were acquired from the NCI, Bethesda, United States of America (USA). All the cells were grown in minimum essential medium (MEM) at RPMI-1640 supplemented with 10% fetal bovine serum (FBS), streptomycin (100 µg/mL), penicillin (100 U/mL), pyruvic acid (0.11 mg/mL), L-glutamine (0.3 mg/mL) and 0.37% NaHCO3. Cells were grown in a 95% air atmosphere and 5% CO2 gas environment at 37 °C with 98% humidity. Cells were treated with EtOH leaf extracts and compound dissolved in DMSO while the untreated cultures received only the vehicle (DMSO, <0.2%, volume percentage).
2.6. Cell viability by MTT assay
The cytotoxicity of the EtOH extract of P. acerifolium leaf and the isolated compound was analyzed by the MTT assay. Human cancer cell lines include breast, colon, the lung was seeded at a density of 6 × 103 cell/200 µL of the medium in 96 well plates. All cancer cells (breast, colon and lung) were treated with leaf extract and compounds for 48 h at different concentrations. The MTT formazon blue crystal was dissolved in 150 µL of DMSO. Finally, absorbance was measured with an ELISA reader at wavelength 570 nm (620 nm for reference wavelength). Percent (%) viability of cell growth was calculated when compares absorbance of treated versus untreated cells. The 50% inhibitory concentration of leaf extract and the compound was defined result, 50% loss of cell viability relative to untreated cells. Doxorubicin was used as a positive control, which was calculated in nmol/L.
2.7. Molecular docking
Molecular docking of a compound with kinases was performed using Autodock Vina (Trott and Olson, 2010) and MGL tools (Sanner, 1999, Morris et al., 2009). The 3D crystallographic structure of PI3K (PDB: 4L23) and mTOR (PDB: 4JT6) were downloaded from the protein data bank (http://www.pdb.rcsb.org) and the isolated compound was drawn using chem3D pro 12.0.2.1076. Water and ligand were removed from kinases (PI3K and mTOR) by using the software Discovery Studio Visualizer. Polar hydrogen atoms and Kollman charges were added to the 3D structure of the protein and saved prepared protein file in a format by Auto Dock Tools-1.5.6. A grid box for PI3K (4L23) was performed with a dimension of 98 × 116 × 126 (X × Y × Z) with grid spacing 1Ao and grid center X = Y = Z = 0.028. A grid box on the active site of mTOR (4JT6) was performed at grid center X = Y = Z = 0.028 with dimension 126 × 126 × 126 (X × Y × Z) and spacing 1Ao. Finally, molecular docking was done by Auto dock vina. One drug (Dactolisib) was screened with PI3K and mTOR kinases for known binding affinity. This drug is used in inhibitory activity of PI3K and mTOR kinases, utilized as a positive control.
3. Results and discussion
3.1. Structure of (3β)-D-friedoolean-14-en-3ol (taraxerol)
The white crystal of the compound was obtained. The FT-IR (KBr) spectrum was displayed an absorbance peak at 3484 cm−1 (stretching vibration of OH bond), 2935 and 2862 (sp3 H-stretching), 1643 (C = C stretching), 1467 and 1382 (sp3 H-bending), 1036 (C-O bending). 1HNMR spectrum of compound in CDCl3 was showing doublet–doublet at δH 3.19 (1H, dd, H-3; CH), one double bond proton at δH 5.53 (1H, dd, H-15; CH) and eight methyl groups (δH 0.98, 0.80, 0.92, 0.82, 1.09, 0.90, 0.95, 0.90; 3H × 8, s). 13C and DEPT-135 NMR spectra was showed eight methyl carbons (δC: 28.0, 15.43, 15.46, 29.83, 25.91, 29.93, 33.35, 21.32), ten methylene carbons (δC: 37.71, 27.13, 18.79, 41.33, 17.50, 35.18, 37.73, 36.68, 33.69, 33.09), four methine carbons (δC: 79.19, 55.96, 48.73, 49.27), six quaternary carbons (δC: 38.97, 37.99, 38.76, 37.56, 158.08, 35.79, 28.80). HSQC was displayed proton- carbon correlation, methyl hydrogens at δH 0.90 in HSQC was showing a correlation with carbon δC 21.32 and in HMBC methyl hydrogens (δH 0.90) was displayed correlation with methyl carbon δC 33.35 and quaternary carbon at δC 28.80. It was indicated that the methyl group (δC 21.32) was connected with methyl carbon (δC 33.35) and quaternary carbon (δC 28.80). Methine proton at δH 1.42 in HSQC was correlated with methine carbon (δC 49.27) and in HMBC (Fig. 1C) methine carbon (δC 49.27) was correlated with two methyl carbon at δC 29.83, 29.93 and methylene carbon at δC 37.73.It was validated that the methine group (δC 49.27) was connected with two methyl carbon (δC 29.83, 29.93) and one methylene carbon (δC 37.73). In HSQC spectrum, methine proton at δH 0.95 was correlated with methine carbon (δC 48.73) and in HMBC methine carbon (δC 48.73) was correlated two methyl carbon δC 25.91, 15.46 and one methylene carbon δC 41.33. It was indicated that the methine group (δC 48.73) was connected with two methyl carbon δC 25.91, 15.46 and one methylene carbon δC 41.33. Methine proton at δH 0.78 in HSQC was correlated with methine carbon δC 55.96 and in HMBC methine carbon was correlated with three methyl carbon δC 28.0, 15.43, 15.46 and one methylene carbon δC 41.33. It was indicated that methine carbon (δC 55.96) was connected to three methyl carbon (δC 28.0, 15.43, 15.46) and one methylene carbon (δC 41.33). Cross peak in COSY (Fig. 1B) spectrum were showed a correlation between H-3 proton (δH 3.19) & H-2 (δH 1.58) and H-5 (δH 0.78) & H-6 (δH 1.58) and H-9 (δH 0.95) & H-11 (δH 1.65) and H-15 (δH 5.5) & H-16 (δH 1.62) (Fig. 1).
Fig. 1.
Structure (A), key 1H-1HCOSY (B) and HMBC (C) correlations of taraxerol.
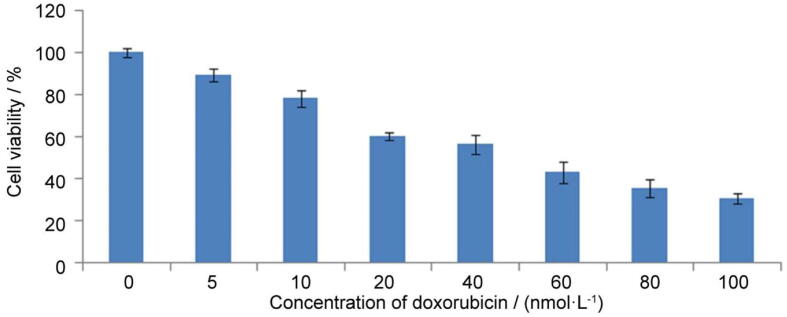
3.2. Cytotoxicity of doxorubicin drug
Cytotoxicity of doxorubicin evaluated on cancer cell line MDA-MB-231 by MTT assay for 48 h. At 5 nmol/L concentration, cell viability of doxorubicin compound reached 10% in MDA-MB-231 cells. After increaseing concentration, cell viability of drug was increased, which was summarized in Fig. 2. At 10 and 20 nmol/L concentration, cell viability reached 25% and 40%, respectively. Inhibition concentration (IC50) of doxorubicin was found at 52 nmol/L in MDA-MB-231 cancer cells. Doxorubin drug is high potent in treatment of cancer, which used as a positive control.
Fig. 2.
Effect of doxorubicin on cell viability of MDA-MB-231 cancer cell at different concentrations for 48 h by MTT assay.
3.3. Cytotoxicity of leaf extract
In vitro, cytotoxicity of the EtOH leaf extract was analyzed against human breast (MDA-MB-231 & BT-549), colon (SW-480) and lung (A-549) cancer cell lines by MTT assay for 48 h. IC50 values (concentration for 50% cell death) are summarized (Fig. 3A). MDA-MB-231 was showing IC50 values at very low concentration (80 µg/mL) which indicates excellent inhibitory activity in the breast cancer cell. Cancer cell line BT-549 was displayed IC50 at low concentration110 µg/mL which reveal a good activity. A-549 and SW-480 were showing the inhibitory effect at concentration 160 µg/mL and 130 µg/mL. From the above investigation, it indicated that the EtOH extract displayed a high inhibitory effect on MDA-MB-231 and BT-549 cell lines.
Fig. 3.
Effects of EtOH leaf extract of P. acerifolium (A) and taraxerol compound (B) on cell viability at different concentrations in breast (MDA-MB-231 and BT-549), colon (SW-480) and lung (A-549) cancer cell lines by MTT assay for 48 h.
3.4. Cytotoxicity of taraxerol
In vitro, the effects of cytotoxicity for taraxerol were evaluated against four human cancer cell lines including MDA-MB-231, BT-549, A-549, and SW-480 for 48 h by MTT based cell viability. IC50 values are listed in Fig. 3B. Taraxerol showed the most potent activity at concentration 160 µg/mL against MDA-MB-231. SW-480 was displayed cytotoxicity at concentration 210 µg/mL which reveal good inhibitory effect. Cancer cells BT-549 and A-549 were displaying cytotoxicity effect at concentration 270 µg/mL and 290 µg/mL which reveal low inhibition activity. The above observation indicated that taraxerol showed good inhibitory activity on the MDA-MB-231 cancer cell line compared with BT-549, A-549 and SW-480.
The extract showed more cytotoxic activity on all cancer cell lines comparing with taraxerol. The extract and taraxerol all displayed excellent inhibitory activity on cancer cell MDA-MB-231.
3.5. Theoretical investigation studies (molecular docking)
The cancer is a main problem of the whole world and patients are being increased day by day. Various drugs have been developed to treat cancer. Dactolisib (BEZ235) is a highly potent drug, which used in dual PI3K/mTOR inhibitors. Dactolisib combined with Everolimus which used 200, 400, and 800 mg dose daily in patients clinically for 28 d (Wise-Draper et al., 2017). The dual PI3K/mTOR inhibitor BEZ235 suppressed tumor growth in vitro and in vivo (Wu et al., 2019). Combination of Dactolisib with Diosmin is effective on HCT-116 colorectal cancer cell line through inhibition of the PI3K/mTOR (Helmy et al., 2020). Dactolisib is a dual PI3K-mTOR inhibitor, which is used in phase I and phase II clinical trials (Yang et al., 2019). We screened dactolisib drug with PI3K and mTOR kinases, found binding energies −11.2 kcal/mole (Table 1, entry 1) and 10.8 kcal/mole (Table 1, entry 2), which used as a positive control.
Table 1.
Optimization studies of a taraxerol compound.
| Entry | Proteins/Drugs | PDB (I.D)a | Docking score /(kcal·mol−1) |
|---|---|---|---|
| 1 | Dactolisib + PI3K | 4L23 | −11.2 |
| 2 | Dactolisib + mTOR | 4JT6 | −10.8 |
| 3 | Taraxerol + P53 | 4HJE | −8.6 |
| 4 | Taraxerol + EGFR | 2JIT | −8.3 |
| 5 | Taraxerol + IGFR | 1IGR | −5.7 |
| 6 | Taraxerol + PDK | 2R7B | −7.7 |
| 7 | Taraxerol + Tankyrase | 5JU5 | −8.2 |
| 8 | Taraxerol + mTOR | 4JT6 | −10.0 |
| 9 | Taraxerol + PTEN | 1D5R | −6.5 |
| 10 | Taraxerol + KRAS | 6N2J | −7.2 |
| 11 | Taraxerol + PI3K | 4L23 | −9.8 |
Obtained from http://www.pdb.rcsb.org.
We investigate taraxerol compound with various proteins, which used in several cancer signaling pathways. P53 is a tumor suppressor protein, which involved in controlling pathways apoptosis, cell cycle (Klein & Vassilev, 2004, Lane, 1992). Epidermal growth factor (EGF) and epidermal growth factor receptor (EGFR) is overexpressed in different types of human cancers (Seshacharyulu et al., 2012). We observed binding energies of the isolated compound with P53 −8.6 kcal/mole (Table 1, entry 3) and with EGFR −8.3 kcal/mole (Table 1, entry 4). The IGF (insulin-like growth factor) play an important role in growth, development, and maintenance of several tissues in the human body (Denduluri et al., 2015). PIP3 converts into PIP2, this, process control by PTEN. We found docking results of the isolated compound with EGFR protein −5.7 kcal/mole (Table 1, entry 5) and with PTEN −6.5 kcal/mole (Table 1, entry 9). PI3Ks play important role in cellular functions as cell growth, survival, differentiation, proliferation. mTOR is a main component of the PI3K/AKT signaling pathway, which regulates cell proliferation, migration, metabolism and survival. We observed best binding interaction of the isolated compound with mTOR −10.0 kcal/mole (Table 1, entry 8) and binding energy of PI3K (Table 1, entry 11) is very close to mTOR. The optimization results indicate that the isolated compound plays a key role in inhibiting PI3K and mTOR. PI3K/AKT/mTOR signaling pathway was showed in Fig. 4. The dual inhibitor compound is best for chemotherapies which can inhibit of the whole pathway.
Fig. 4.
Taraxerol targeting of PI3K/AKT/mTOR pathway.
3.6. Molecular docking analysis with PI3K
Taraxerol was screened with PI3K in silico, which indicates a favorable binding with docking score −9.8 Kcal/mol. PI3K was displays important hydrogen bond, hydrophobic interaction (Fig. 5) with residues TYR, PRO, LYS, and THR. The binding interaction of taraxerol was investigated with PI3K, which indicates a non-classical hydrogen bond interaction of THR-679 at 3.32 distance with C-3 carbon. The taraxerol binding investigations were focused on alkyl hydrophobic and mixed pi/alkyl hydrophobic interaction of LYS-640 at 5.35 distance with C-23, TRY-470 at 5.08 distance with C-29, PRO-447 at 3 × 4.31, 4.89 and 5.0 distance with C-27 and C-30 and PRO-449 at 3.48, 4.89, 5.09, and 5.25 distance with C-25.
Fig. 5.
Binding studies of taraxerol (ball and stick) with PI3K kinases (4L23) (A), hydrogen and hydrophobic interaction with amino acid PRO, TYR, LYS, THR (B), binding distance of PI3K kinases (C), group of ligand used in binding (D).
3.7. Molecular docking analysis with mTOR
In silico, the isolated compound was docked successfully with mTOR, which indicating best binding with affinity −10 kcal/mol. Hydrophobic interaction (Fig. 6) was displayed with amino acid PRO, LEU, ALA, and ILE. The binding interaction of taraxerol was analyzed with mTOR, which indicates alkyl hydrophobic interactions of PRO-1975 at 4.64 distance with C-30, ILE-1939 at 4.40 distance with C-27, ALA-1971 at 3.46, 4.49 distance with C-27, LEU-1936 at 3.94, 4.07, 4.71, 5.0, 5.28 distance with C-25 and C-27.
Fig. 6.
Interaction of taraxerol with mTOR (A), group of ligand used in hydrophobic interaction (B), binding interaction of ligand with PRO, LEU, ALA and ILE (C), and binding distance of amino acid with ligand (D).
4. Conclusion
In summary, we have isolated taraxerol compound from the leaf extract of P. acerifolium and structure was confirmed by various spectroscopic techniques. Cytotoxicity of the extract and compound were screened on MDA-MB-231, BT-549, A-549, and SW-480 cancer cell lines. The extract and the compound were exhibited best result on MDA-MB-231 cancer cell line. The molecular docking also indicated the best results with PI3K (-9.8 Kcal/mol) and mTOR (-10.0 Kcal/mol). Thus extract and the compound may be useful for the treatment of cancer.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are thankful to UGC, New Delhi, India for institutional fellowship. The authors are also grateful to the Chairman, Department of Chemistry, AMU, Aligarh, for providing research facilities.
References
- Balachandran P., Govindarajan R. Cancer—an ayurvedic perspective. Pharmacological Research. 2005;51(1):19–30. doi: 10.1016/j.phrs.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Caius J.F. The medicinal and poisonous plants of India. Indian Medicinal Plants. 1990;2:489. [Google Scholar]
- Cantley, L. C. (2002). The phosphoinositide 3-kinase pathway. Science, 296, 1655-1657. [DOI] [PubMed]
- Ediriweera M.K., Tennekoon K.H., Samarakoon S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Seminars in Cancer Biology. 2019;59:147–160. doi: 10.1016/j.semcancer.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Denduluri S.K., Idowu O., Wang Z., Liao Z., Yan Z., Mohammed M.K.…Luu H.H. Insulin-like growth factor (IGF) signaling intumorigenesis and the development ofcancer drug resistance. Genes & Diseases. 2015;2(1):13–25. doi: 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J.A., Luo J.i., Cantley L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nature Reviews Genetics. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Kirtikar K.R., Basu B.D. 2nd ed. Allahabad. In Indian Medicinal Plants Allahabad; Lalit Mohan Basu: 1933. In Indian Medicinal Plants; p. 374. [Google Scholar]
- Gupta P.C., Bishnoi P. Structure of new acid polysaccharide from the bark of Pterospermum acerifolium. Journal of the Chemical Society Perkin Transactions. 1979;1(7):1680–1683. [Google Scholar]
- Gupta P.C., Suresh C., Rizvi S. Chemical examination of the flower of Pterospermum acerifolium. Planta Medica. 1972;21:358–363. doi: 10.1055/s-0028-1099565. [DOI] [PubMed] [Google Scholar]
- Haar E.V., Lee S.I., Bandhakavi S., Griffin T.J., Kim D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nature Cell Biology. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K.I., Oshiro N., Hidayat S., Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Helmy M.W., Ghoneim A.I., Katary M.A., Elmahdy R.K. The synergistic anti–proliferative efect of the combination of diosmin and BEZ–235 (dactolisib) on the HCT–116 colorectal cancer cell line occurs through inhibition of the PI3K/Akt/mTOR/NF–κB axis. Molecular Biology Reports. 2020;47:2217–2230. doi: 10.1007/s11033-020-05327-4. [DOI] [PubMed] [Google Scholar]
- Klein C., Vassilev L.T. Targeting the p53–MDM2 interaction to treat cancer. British Journal of Cancer. 2004;91(8):1415–1419. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.P. Guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Liu P., Cheng H., Roberts T.M., Zhao J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nature Reviews Drug Discovery. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna A.K., Jena J., Manna S. The antiulcer activity of Pterospermum acerifolium barks extract in experi-mental animal. Journal of Pharmacy Research. 2009;2:785–788. [Google Scholar]
- Manna A.K., Jena J. Anti inflammatory and analgesic activity of bark extract of P. acerifolium. International Journal of Current Pharmaceutical Research. 2009;1:32–37. [Google Scholar]
- Manna A.K., Manna S., Behera A.K. In vitro antioxidant activity of P. acerifolium barks. Journal of Pharmacy Research. 2009;2:1042–1044. [Google Scholar]
- Manning, B. D., (2004). Balancing Akt with S6K: Implications for both metabolic diseases and tumorigenesis. Journal of Cell Biology, 167, 399-403. [DOI] [PMC free article] [PubMed]
- Morris G.M., Huey R., Lindstrom W. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. Journal of Computational Chemistry. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan A.K. mTOR: Role in cancer, metastasis and drug resistance. Seminars in Cancer Biology. 2019;59:92–111. doi: 10.1016/j.semcancer.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Peng T., Golub T.R., Sabatini D.M. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Molecular & Cellular Biology. 2002;22(15):5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud, C. G. (2009). mTORC1 signalling and mRNA translation. Biochemical Society Transactions, 37, 227-231. [DOI] [PubMed]
- Rizvi S., Sultana J. Phytochemical studies of the flower of Pterospermum acerifolium. Phytochemistry. 1972;11:856–858. [Google Scholar]
- Smith A.L., D’Angelo N.D., Bo Y.Y., Booker S.K., Cee V.J., Herberich B., Norman M.H. Structure-based design of a novel series of potent, selective inhibitors of the class I phosphatidylinositol 3-kinases. Journal of Medicinal Chemistry. 2012;55(11):5188–5219. doi: 10.1021/jm300184s. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Thoreen C.C., Peterson T.R., Lindquist R.A., Kang S.A., Spooner E., Sabatini D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Molecular Cell. 2007;25(6):903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sanner M.F. Python a programming language for software integration and development. Journal of Molecular Graphics and Modelling. 1999;17:57–61. [PubMed] [Google Scholar]
- Shaw R.J., Cantley L.C. Ras, PI3K and mTOR signaling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Kim D.-H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Sabatini D.M. Rictor, a novel vinding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current Biology. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Seshacharyulu P., Ponnusamy M.P., Haridas D., Jain M., Ganti A.K., Batra S.K. Targeting the EGFR signaling pathway in cancer therapy. Expert Opinion on Therapeutic Targets. 2012;16(1):15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I., Sawyers C.L. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nature Reviews Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wise-Draper T.M., Moorthy G., Salkeni M.A., Karim N.A., Thomas H.E., Mercer C.A.…Morris J.C. A phase 1b study of the dual PI3K/mTOR inhibitor dactolisib (BEZ235) combined with everolimus in patients with advanced solid malignancies. Targeted Oncology. 2017;12:323–332. doi: 10.1007/s11523-017-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.Y., Wu H.C., Wu J.E., Huang K.Y., Yang S.C., Chen S.X., Hong T.M. The dual PI3K/mTOR inhibitor BEZ235 restricts the growth of lung cancer tumors regardless of EGFR status, as a potent accompanist in combined therapeutic regimens. Journal of Experimental Clincal Cancer Research. 2019;38(1) doi: 10.1186/s13046-019-1282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Nie J.i., Ma X., Wei Y., Peng Y., Wei X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Molecular Cancer. 2019;18(1) doi: 10.1186/s12943-019-0954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]