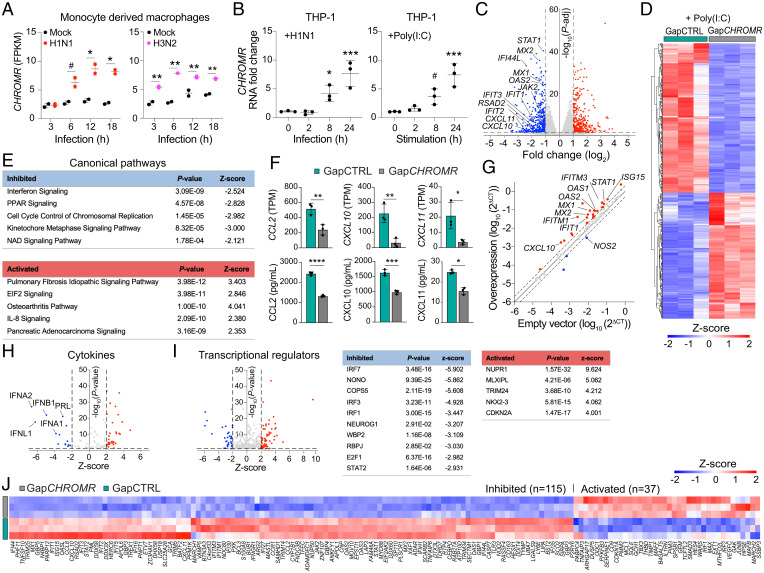
Fig. 2.
CHROMR deficiency leads to diminished expression of ISGs. (A) Time course of CHROMR expression (FPKM) in human monocyte-derived macrophages infected with A/California/04/09 (H1N1) virus, A/Wyoming/03/03 (H3N2) virus, or mock infected. (B) qPCR analysis of CHROMR in human THP-1 macrophages infected with A/WSN/1933 (H1N1) virus(1,000 PFU) or stimulated with synthetic dsRNA poly(I:C) (1 μg/mL). (C) Volcano plot showing differentially expressed genes in CHROMR-depleted (GapCHROMR-treated) and control (GapCTRL-treated) THP-1 macrophages after poly(I:C) (1 μg/mL, 8 h) stimulation and RNA-seq. Dashed lines indicate fold change (log2) = ±1; P-adj = 0.05; red dots indicate up-regulated genes; blue dots indicate down-regulated genes; gray dots indicate nonsignificantly changed genes. (D) Hierarchical clustering heatmap showing normalized gene expression values in THP-1 macrophages treated with GapCHROMR or GapCTRL in poly(I:C) stimulated conditions (1 μg/mL, 8h). Cutoffs used for visualization: −2 < fold change > 2; and P-adj < 0.05. (E) List of most affected canonical pathways identified through Ingenuity Pathway Analysis of (C) ranked by P-adj. (F) Expression of top chemokine genes differentially regulated in CHROMR-depleted and control THP-1 macrophages. Top row: RNA-seq normalized expression counts (TPM) after poly(I:C) (1 μg/mL, 8h). Bottom row: Immunoassay of protein levels after poly(I:C) (1 μg/mL, 24h). (G) Gene expression profiling of 84 interferon-stimulated genes in THP-1 macrophages stably overexpressing CHROMR or an empty vector control. Up-regulated genes are indicated in red and down-regulated genes in blue. Genes indicated are P < 0.1. (H and I) Predicted cytokine (H) and transcriptional regulators (I) of differentially expressed genes in (C); dashed lines indicate Z-score = ±2 and P-adj = 0.05; red and blue dots indicate significantly up-regulated and down-regulated factors, respectively. (J) Hierarchical clustering heatmap showing Z-scores of differentially expressed ISGs in CHROMR-depleted and control THP-1 macrophages treated with poly(I:C) (1 μg/mL) for 8h, P-adj < 0.05. Data are mean ± SEM for 2 (A), 3 (B–F [Top], G–J) independent experiments, or representative of 3 independent experiments (F [Bottom]). P values were calculated via repeated measures two-way ANOVA with Sidak’s multiple comparison test (A), one-way ANOVA with Dunnett’s multiple comparison test (B), right-tailed Fisher’s exact test (E, H, and I), or two-tailed unpaired Student’s t test (F and G). #P < 0.1; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.