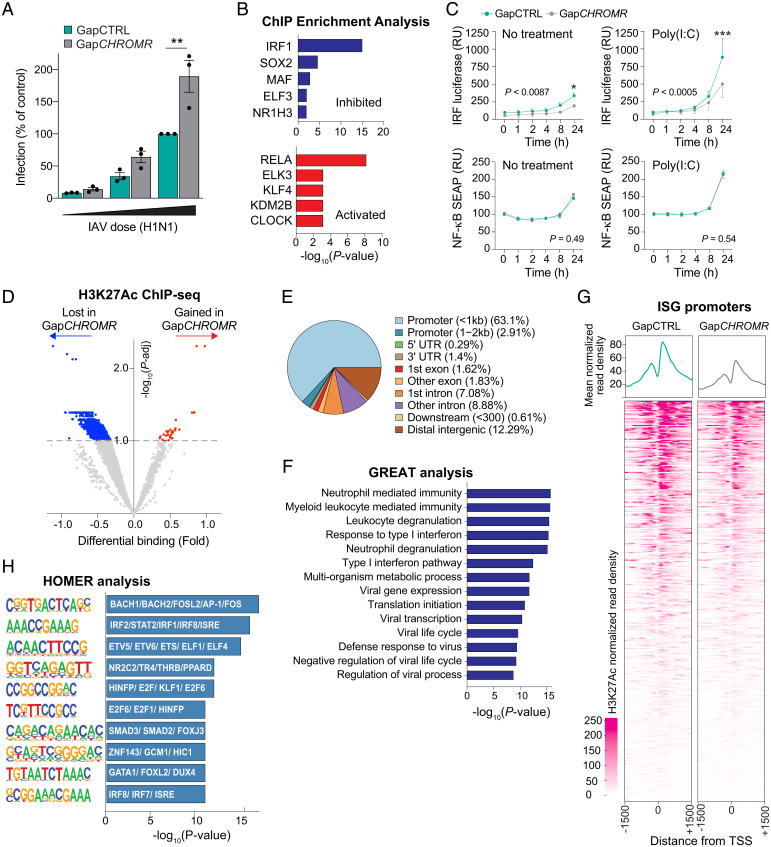
Fig. 3.
CHROMR is required to restrict influenza virus and activate ISG transcription. (A) Percentage of viral infection in CHROMR-depleted (GapCHROMR-treated) and control (GapCTRL-treated) THP-1 macrophages challenged with A/WSN/1933 (H1N1) virus at increasing doses (100, 500 or 1,000 PFU). Percentages were calculated relative to GapCTRL transfection at highest infection rate. (B) Transcription factor binding enrichment scores for ISGs differentially expressed in CHROMR-depleted and control THP-1 macrophages stimulated with poly(I:C) via ChIP enrichment analysis (ChEA 2016) database gene set library. (C) Reporter assay for IRF-driven transcription (Top: luciferase; RU, relative units) or NF-κB–driven transcription (Bottom: SEAP, secreted alkaline phosphatase) in THP-1 Dual Reporter macrophages transfected with GapCHROMR or GapCTRL and left untreated or stimulated with poly(I:C) (1 μg/mL). Relative expression is normalized to time 0 (=100). (D) Volcano plot showing differential H3K27Ac modification in CHROMR-depleted and control THP-1 macrophages stimulated with poly(I:C). ChIP-seq reads that are gained or lost after CHROMR knockdown are indicated in red and blue, respectively. Dashed line indicates P-adj < 0.1. (E) Genomic distribution of H3K27Ac marks lost after CHROMR knockdown identified in D, P-adj < 0.1. UTR, untranslated region. (F) List of biological processes identified via Genomic Regions Enrichment Annotations Tool (GREAT) analysis of H3K27Ac-depleted promoter regions. (G) Metagene plots showing the mean (Top) and individual unique positions (Bottom) of normalized H3K27Ac read density around the transcription start site (TSS ± 1,500 base pairs) of ISGs in THP-1 macrophages transfected with GapCHROMR or GapCTRL. (H) Hypergeometric Optimization of Motif EnRichment (HOMER) analysis of promoter regions depleted of H3K27Ac after CHROMR knockdown, showing transcription factors with highest similarity score in motif indicated in bars. Data are mean ± SEM for three independent experiments. P values were calculated via repeated measures two-way ANOVA with Sidak’s multiple comparison test (A and C) or binomial test (B, F, and H). *P ≤ 0.05; **P < 0.01; ***P < 0.001.