Fig. 2.
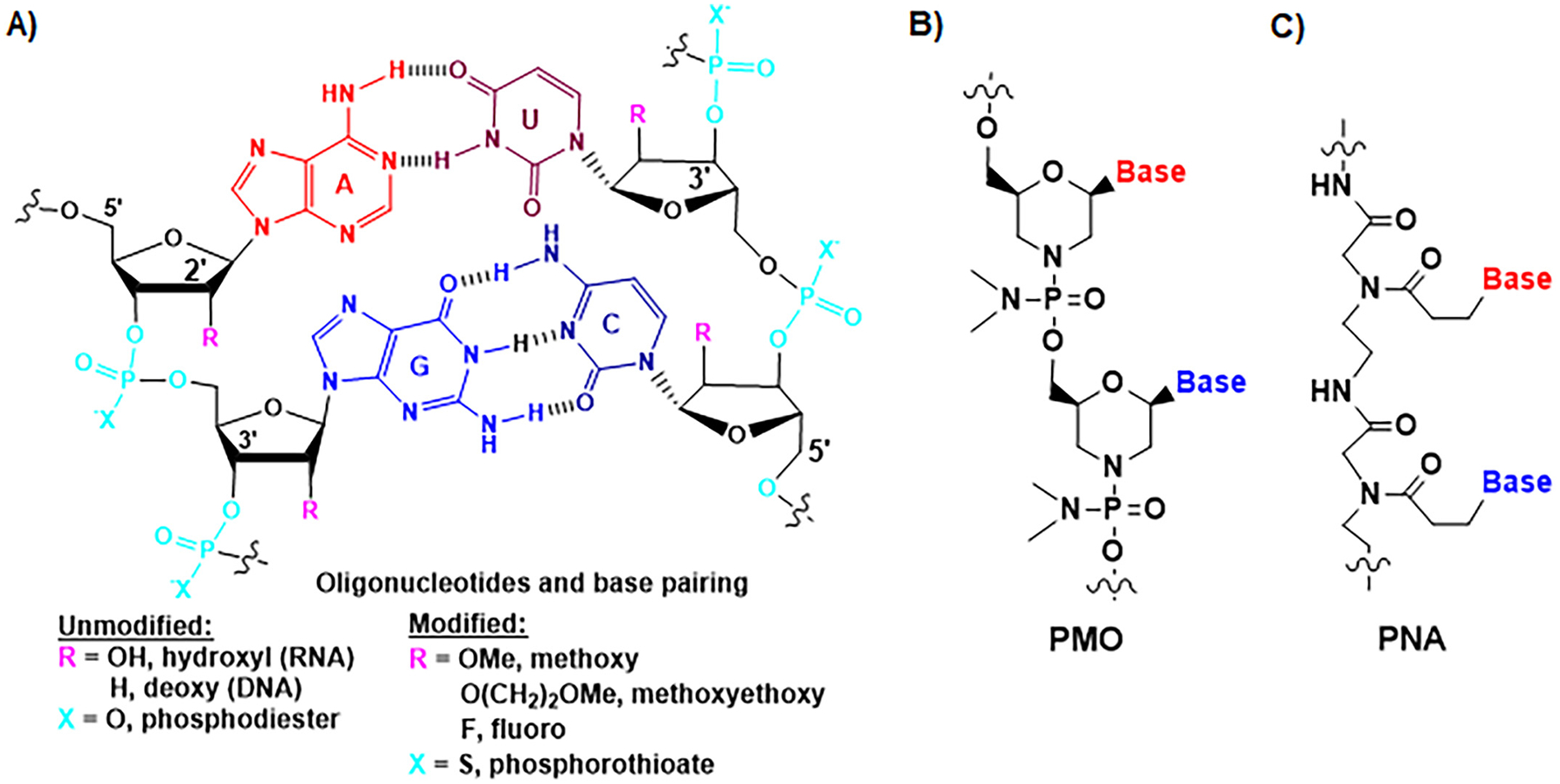
Chemical structures of natural and synthetic oligonucleotides. (A) The nucleobases are paired through hydrogen bonds (H-bonds). Adenine (A) and uracil (U) (or thymine (T) or 5-methyluracil (m5U)) pairs involve two H-bonds, and guanine (G) and cytosine (C) pairs consist of three H-bonds. The phosphodiester bond linkage and 2′-hydroxyl group on ribose ring within natural RNA molecules are two “soft spots” that are commonly modified as other forms towards a greater metabolic stability, such as phosphorothioate, 2′-methoxy, 2′-methoxyethoxy, and 2′-fluoro. (B) Synthetic phosphorodiamidate morpholine oligonucleotide (PMO) or morpholino oligomer is comprised of advanced modifications, in which a six-membered morpholine replaces the five-membered ribose ring and phosphoramidate substitutes the phosphodiester linkage while nucleobases are retained for pairings. (C) Peptide nucleic acid (PNA) is another type of artificial oligonucleotide in which nucleobases are connected with N-(2-aminoethyl)-glycine units linked by peptide bonds.
