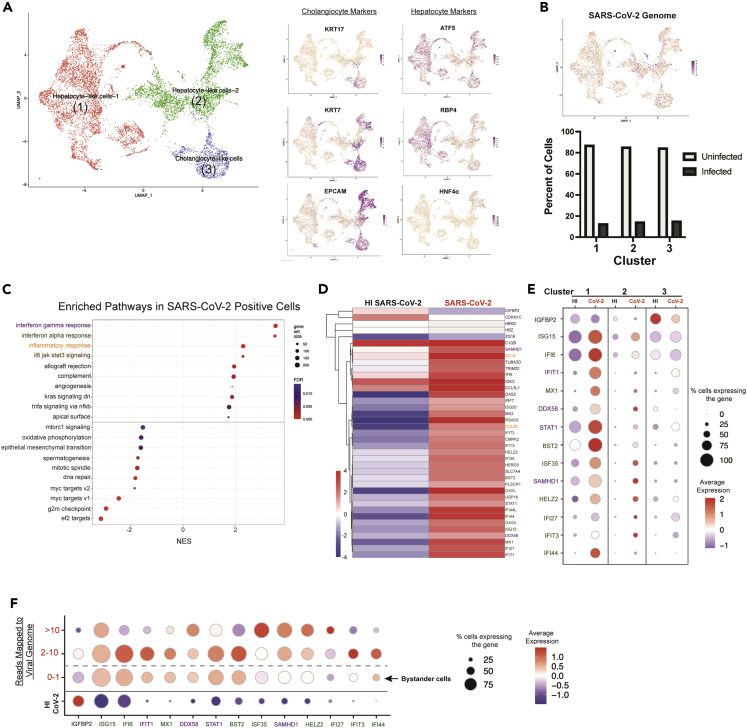
Figure 2.
Transcriptomic response of HLOs to SARS-CoV-2 infection. HLOs were infected with SARS-CoV-2 and sequenced with 10x single-cell sequencing
(A) The three major clusters were identified as (1) Hepatocyte-like cells 1 (ATF5 & RBP4 positive) (2) Hepatocyte-like cells 2 (positive for both hepatocyte and cholangiocyte markers) and (3) Cholangiocyte-like cells (KRT7, KRT17 & EPCAM positive).
(B) Projection of cells with SARS-CoV-2 transcripts onto the UMAP. The graph below shows the percentage of cells with at least one read mapping to the SARS-CoV-2 genome (infected) in each cluster.
(C) Gene set enrichment analysis (GSEA) showing the top upregulated and downregulated gene sets in infected HLO cells versus cells in HLOs exposed to HI SARS-CoV-2.
(D) Heatmap of all significantly differentially expressed genes (Adj. p value < 0.05) in infected HLO cells compared to cells in HLOs exposed to HI SARS-CoV-2. Differential expression was conducted on aggregated counts of all the cells in each condition and included HLOs generated from H1 ESCs and 1016 IPSCs. Color of gene name corresponds to associated gene set (purple = IFNγ, green = IFNα, orange = inflammatory response).
(E) Dot-plot showing expression of top differentially expressed genes from D - that are expressed in at least 50% of one of the conditions plotted-in each major cluster in infected HLO cells versus cells in HLOs exposed to HI SARS-CoV-2.
(F) Expression pattern of top differentially expressed genes showed in E in relation to the number of SARS-CoV-2 genomes detected in the cell. For panels (A–C), data are from HLOs generated from H1 ESCs and 1016 IPSCs. For panels (D–F), data are from HLOs generated from H1 ESCs.