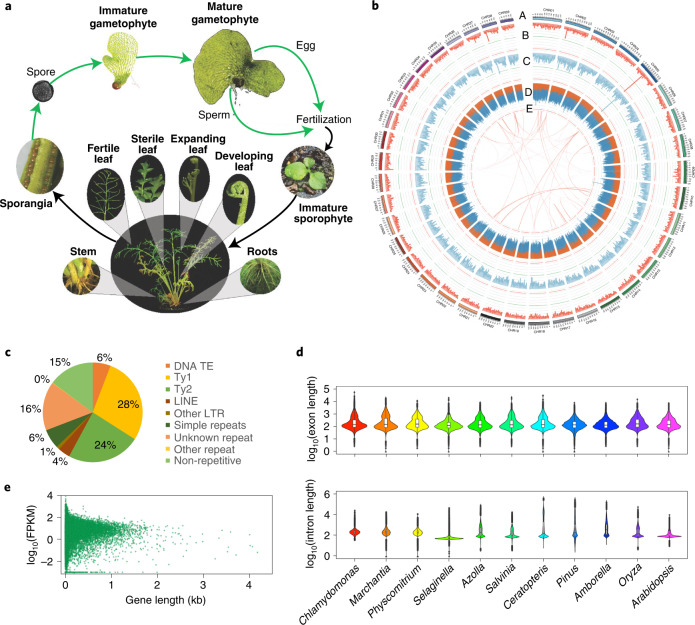
Fig. 1. Ceratopteris richardii life cycle and genome assembly characteristics.
a, Life cycle of Ceratopteris with tissues sampled for RNA-seq in bold. Images are not to scale. b, Genome assembly of Ceratopteris with: (A) chromosomes, (B) gene density in a 3-Mb sliding window, maximum value of 139; (C) mRNA expression density in a 3-Mb sliding window, maximum value of 170; (D) long terminal repeat retrotransposon density in a 3-Mb sliding window, orange and blue bands represent Ty3 and Ty1 LTRs, respectively, maximum value of 970; and (E) intragenomic syntenic regions of ten or more genes. Green horizontal lines represent the 5th percentile, red horizontal lines represent the 95th percentile. c, Genome composition of Ceratopteris. LTR, long terminal repareat; TE, transposable element. d, Intron and exon lengths from a green alga (n = 166,499 exons; 147,269 introns), liverwort (n = 137,019 exons; 112,345 introns), moss (n = 587,902 exons; 501,233 introns), lycophyte (n = 197,720 exons; 162,895 introns), two water ferns (Azolla: n = 127,875 exons; 107,674 introns; Salvinia: n = 122,980 exons; 103,200 introns), Ceratopteris (n = 437,785 exons; 400928 introns), gymnosperm (n = 268,745 exons; 187,344 introns), basal angiosperm (n = 111,241 exons; 83,928 introns), monocot (n = 196,916 exons; 152,273 introns) and eudicot (n = 313,952 exons; 237,746 introns). The widest part of the violin plot represents the highest point density, whereas the top and bottom are the maximum and minimum data respectively. Box plots are in the middle of violin plots, the top and bottom lines represent 25th and 75th percentiles, the centre line is the median and whiskers are the full data range. e, Correlation between fragments per kilobase million (FPKM) and gene length at genome-wide level of Ceratopteris.