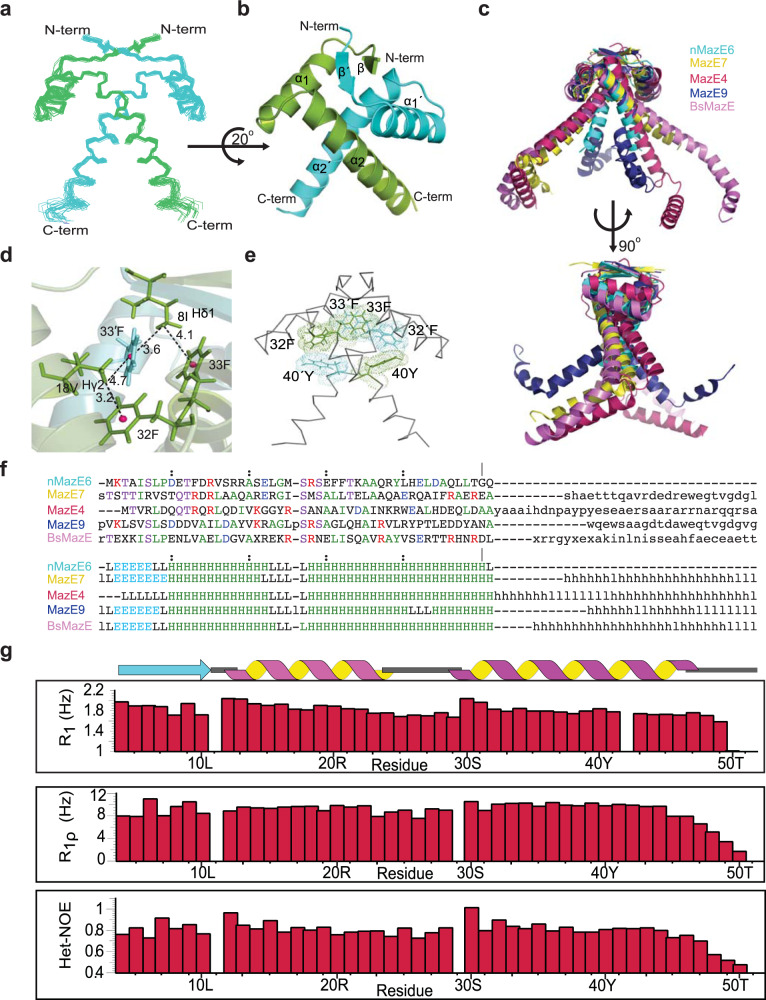
Fig. 3. Quaternary structure and dynamics of nMazE6.
a Ensemble of twenty lowest energy structures calculated for nMazE6 emphasizing the structural symmetry of the dimer. The structures have been superposed on backbone N, Cα and C′ atoms for residues 4–52 and 4′–52′. b Cartoon representation of the lowest energy structure. The secondary structural elements are labeled in order of sequence in each subunit. The figure was generated in PyMol using the structural coordinates of nMazE6. c Alignment of MazE homologs: nMazE6 (cyan), MazE7 (yellow), MazE4 (magenta), MazE9 (navy) and BsMazE (mauve), showing the difference between them. d CH ••• π interactions between 18Vγ and 32F and 8Iδ and 33′ F resulting in upfield shifted lines at −0.15 and −0.18 ppm (Supplementary Fig. S5b) respectively. The distance between the methyl groups and the aromatic ring centroid (pink) is indicated by dashed line. e Aromatic cluster formed by 40Y-32′ F-33F-33′ F-32F-40′ Y. f Structure based sequence alignment of nMazE6 with other MazE from Mtb and Bacillus subtilis MazE. PDB ID: nMazE6 (7WJ0), MazE7(6A6X), MazE4 (5XE3), MazE9 (6KYT) and BsMazE (4ME7) respectively. Structural alignment was done using the DALI server68(g) R1, R1ρ relaxation rates and steady-state heteronuclear {1H} - 15N NOEs for 15N backbone nuclei of nMazE6, plotted against residue number. Source data are provided in the Supplementary Data 1.