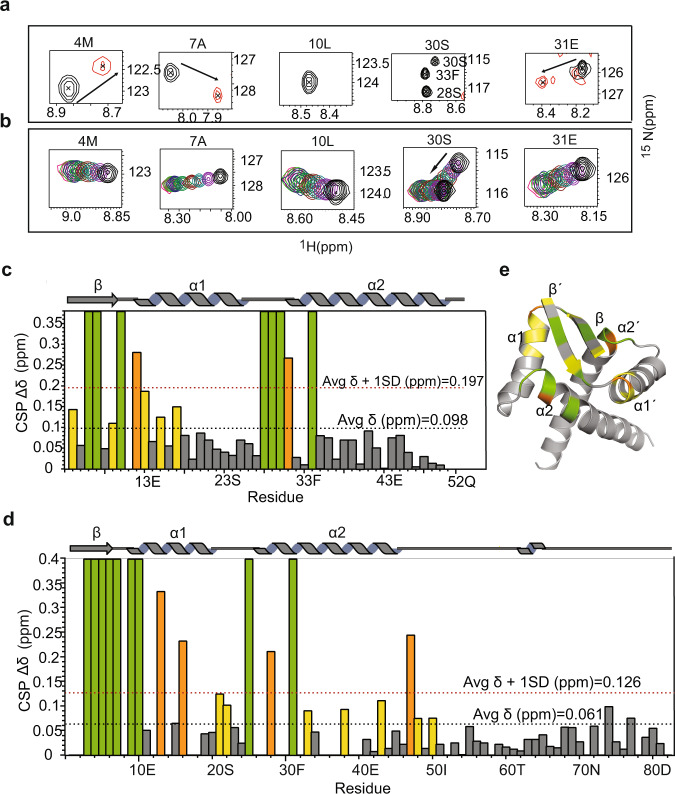
Fig. 5. Chemical Shift Perturbation (CSP) upon operator DNA binding.
a Chemical shift perturbations observed for few residues in the 1H- 15N HSQC (Supplementary Fig. S8b) of nMazE6 upon titration with cognate operator DNA at a 1:2 (nMazE6:DNA) molar ratio. Spectra of free nMazE6 (Black) and nMazE6 in complex with operator DNA at a protein:DNA ratio of 1:2 (Red) are shown. Changes in chemical shifts of backbone amide resonance position between the first and last titration points are indicated by black arrows. For CSP of MazE6, refer Fig. S8A. b Chemical shift perturbations of key residues observed in the 1H- 15N HSQC of nMazE6 upon titration with mazEF6 mutated operator DNA. For full spectrum, refer Supplementary Fig. S9b. c Plot of cumulative CSP as a function of the primary structure of nMazE6. d Plot of cumulative CSP as a function of the primary structure of MazE6. Average CSP and average CSP plus one standard deviation are indicated by dotted lines (Black) and (Red), respectively. e Residues that showed above-average CSP (yellow), above-average CSP + 1SD (orange), and residues that completely broadened and hence disappeared upon DNA binding (green) are mapped on the structure of nMazE6. Source data are provided in the Supplementary Data 1.