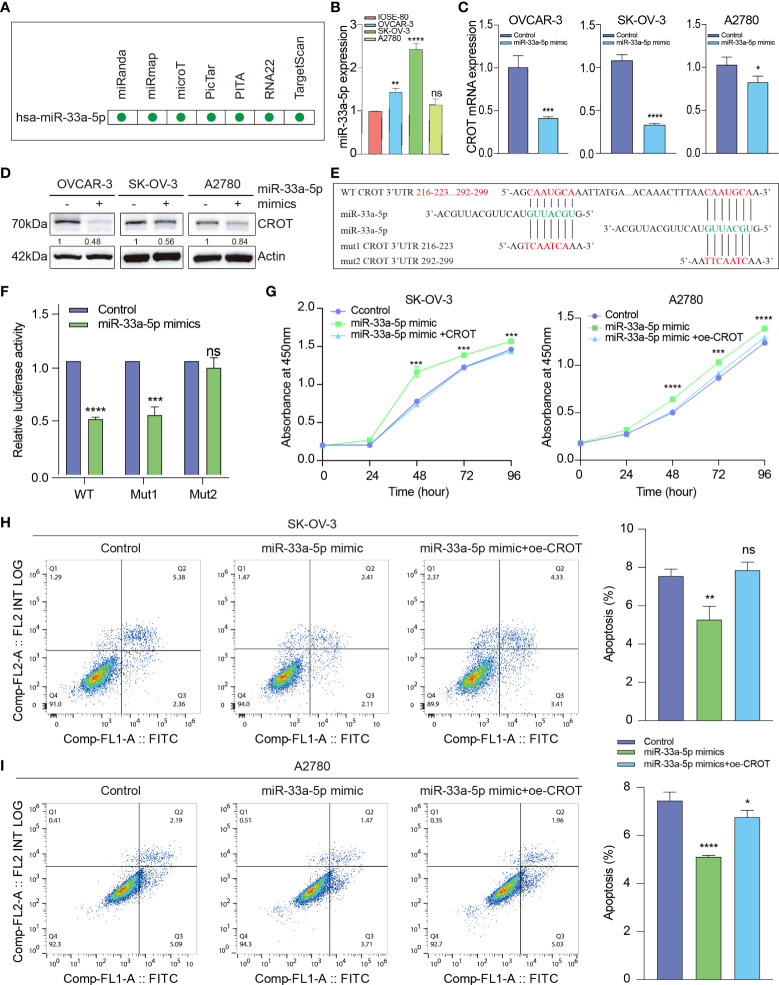
Figure 6.
The function of miR-33a-5p in ovarian cancer cells. (A) Prediction of hsa-miR-33a-5p by the Encyclopedia of RNA Interactomes (ENCORI) online platform, including miRanda, miRmap, microT, PicTar, PITA, RNA22, and TargetScan programs. (B) Detection of the expression of miR-33a-5p in OC cells (OVCAR-3, SK-OV-3, and A2780) and non-tumorous ovarian epithelial cells (IOSE-80) by qRT-PCR. (C, D) Detection of CROT mRNA and protein in OVCAR-3, SK-OV-3, and A2780 cells after the treatment of cells with miR-33a-5p mimics by qRT-PCR and Western blot. (E) Match of two potential binding sites of miR-33a-5p to the 3’UTR of CROT using the TargetScan database. The sequences of wild-type and 2 mutants of the 3’UTR of CROT were shown. (F) Detection of luciferase activity. 293T cells were transfected with luciferase report plasmids (WT, Mut1, or Mut2), followed by the treatment of miR-33a-5p mimics. The luciferase activity was detected by the dual-luciferase assay. (G) Measurement of cell viability. SK-OV-3 and A2780 cells were treated with miR-33a-5p mimics in the presence or absence of CROT-overexpressing plasmids. The cell viability was measured by the CCK-8 assay. (H, I) Detection of apoptotic cells. SK-OV-3 and A2780 cells were treated with miR-33a-5p mimics in the presence or absence of CROT-overexpressing plasmids. Apoptotic cells were detected by flow cytometry. Histograms show the statistical analyses of the data. Assays were repeated at least three times. Data were presented as mean ± SEM. *P <0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant; WT, wild type; Mut1, mutant 1; Mut2, mutant 2; 3’UTR, 3’ untranslated region; oe-CROT, overexpression of CROT.