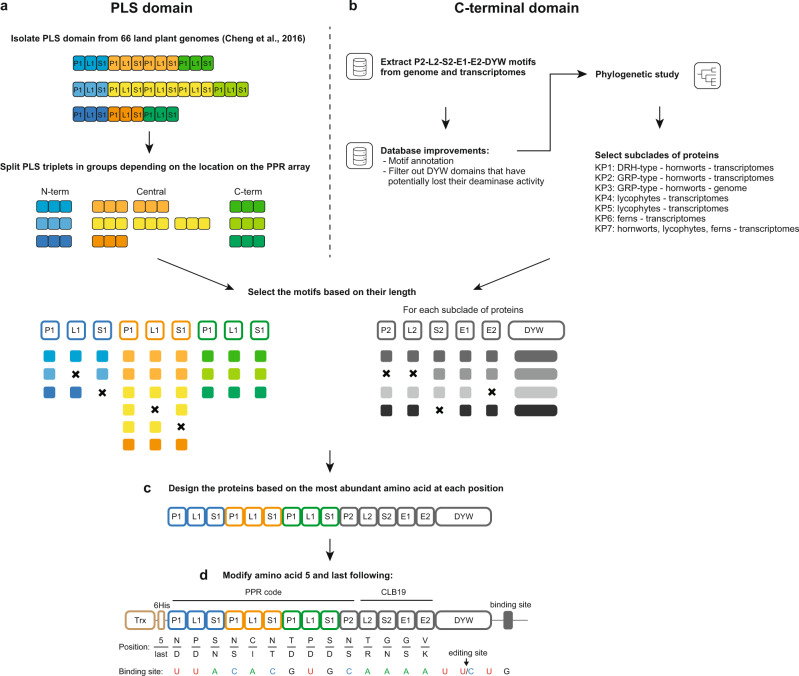
Fig. 1. Schematic depiction of the workflow for designing the PLS and C-terminal domains of designer DYW:KP proteins.
a The P1L1S1 triplets used for the design of the PLS domain were isolated from land plant genomes and selected based on their location in the PLS domain: the ‘N-term’ triplets correspond to the first P1L1S1 triplet in the PLS domain, the ‘Central’ triplets are preceded and followed by a P1L1S1 triplet, and the ‘C-term’ triplets precede a P2L2S2 triplet. b The C-terminal domains were designed on the P2, L2, S2, E1, and E2 PPR-like motifs and DYW domains isolated in hornworts, lycophytes and ferns transcriptomes and Anthoceros angustus genome. After improving the motif database, a phylogenetic analysis was performed to isolate subclades of proteins. c After selecting the PPR motifs on their average length, a unique PLS domain composed of three P1L1S1 triplets and seven C-terminal domains (one for each subclade of proteins identified in (b)) were designed based on consensus sequences. d The amino acids involved in the RNA recognition were mutated to recognize specifically AtrpoA. The DYW:KP proteins overexpressed in Rosetta 2 cells were tagged with an N-terminal thioredoxin (Trx) domain and His-tag (6His). The targeted editing site (AtrpoA) is localized downstream of the stop codon. Below each PPR motif, the two amino acids determining the target specificity are aligned with the AtrpoA editing site. U/C indicates the editing site. Arrows indicate the design flow.