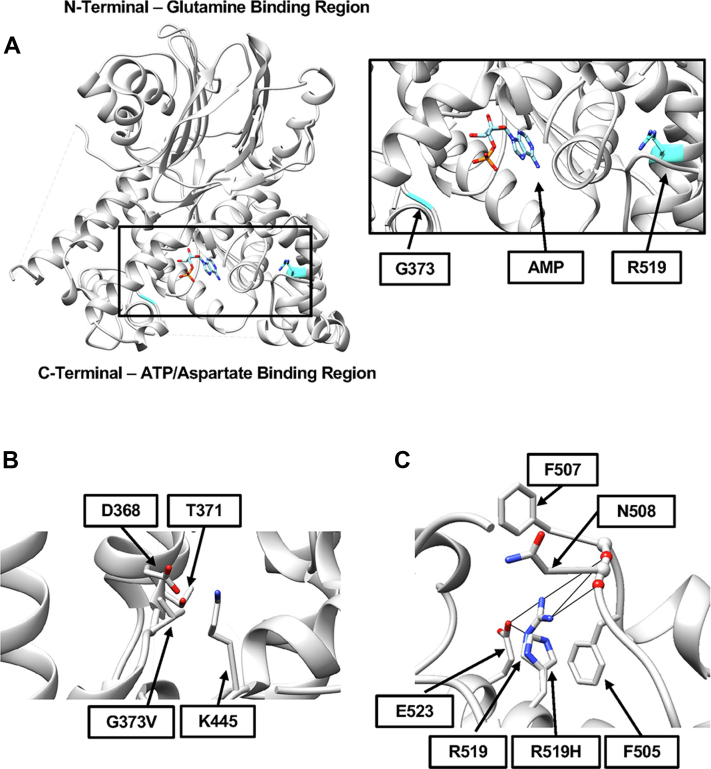
Figure 2.
In silico modeling of variants within the ASNS protein structure.A, the locations of amino acids G373 and R519 (shown in cyan) were modeled using the human ASNS crystal structure (7). The insert shows a close-up of the C-terminal ATP/Asp-binding region illustrating the distance from the AMP-binding site for G373 (a minimum of 8.9 Å) and R519 (a minimum of 24 Å). B, steric clashes are predicted between the most probable G373V rotamer and residues D368, T371, and K445. C, the region surrounding the R519H variant is shown to illustrate the possible loss of hydrogen bonding and possible steric hindrance. For all panels, nitrogen atoms are shown in blue, oxygen atoms in red, and phosphorous in orange. ASNS, asparagine synthetase.