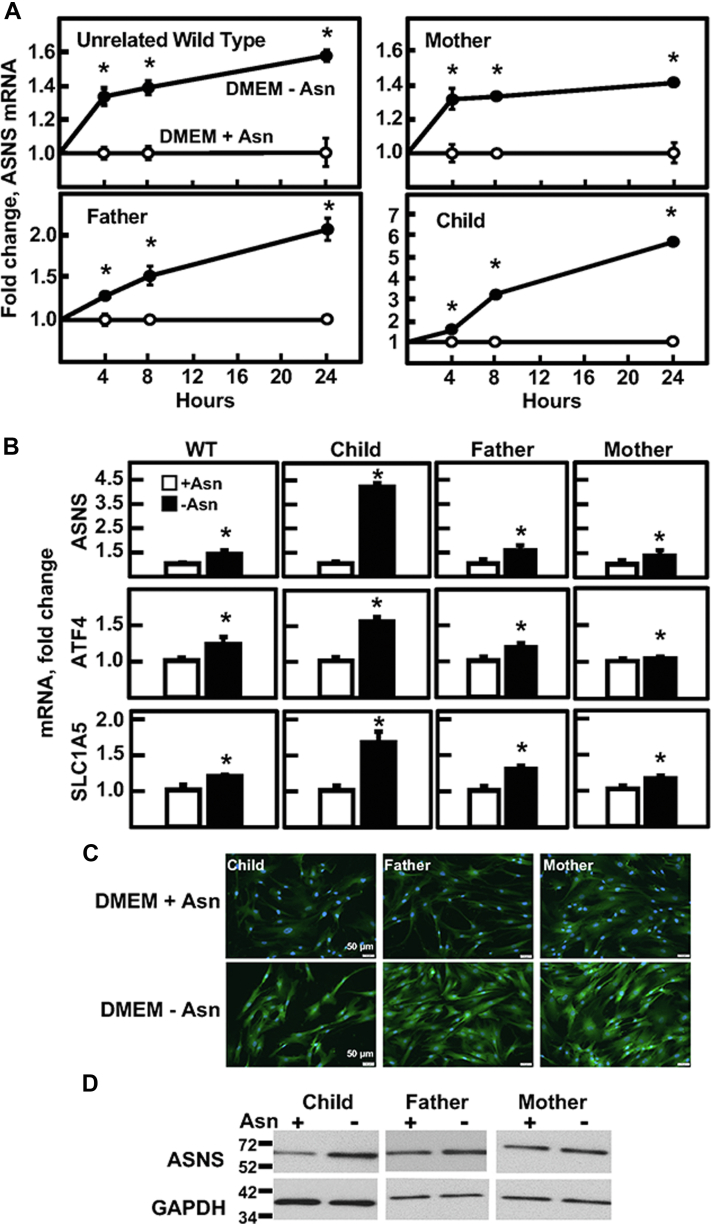
Figure 3.
Amino acid–regulated gene expression in cultured ASNSD fibroblasts. Primary cultures of fibroblasts from the child, both parents, and a nonrelated WT fibroblast cell line were cultured in DMEM ± Asn for 0–24 h. A, ASNS mRNA was measured by qRT-PCR of total RNA and the fold change was calculated for all fibroblast cell lines. The ASNS mRNA values were normalized to the mRNA abundance of GAPDH in the same sample. B, the RNA samples described for panel (A) were analyzed by qRT-PCR for the mRNA abundance of ASNS, ATF4, and the ASCT2 amino acid transporter SLC1A5, each normalized to the GAPDH mRNA content of each sample. For panels (A) and (B), the data are shown as the averages ± SDs for assays in triplicate. Each experiment was repeated at least once with different batches of cells to establish reproducibility. Where not shown, the SD bars are within the symbol. Based on a two-tailed Student’s t test, an asterisk indicates a p value of ≤ 0.05. C, to analyze the uniformity of the ASNS protein content across all cells, immunofluorescence was performed for the fibroblast cell line from each family member after a 24 h incubation in DMEM ± Asn. Immunoblotting was performed for each sample to identify the ASNS protein based on size, with GAPDH used as a control (D). The data shown in panels (C) and (D) are from a representative experiment, and each experiment was repeated at least once to confirm reproducibility. ASNS, asparagine synthetase; ASNSD, ASNS deficiency; qRT-PCR, quatitative real-time PCR.