Figure 2 |. Chromosome segregation phenotypes of RNAi are highly conserved.
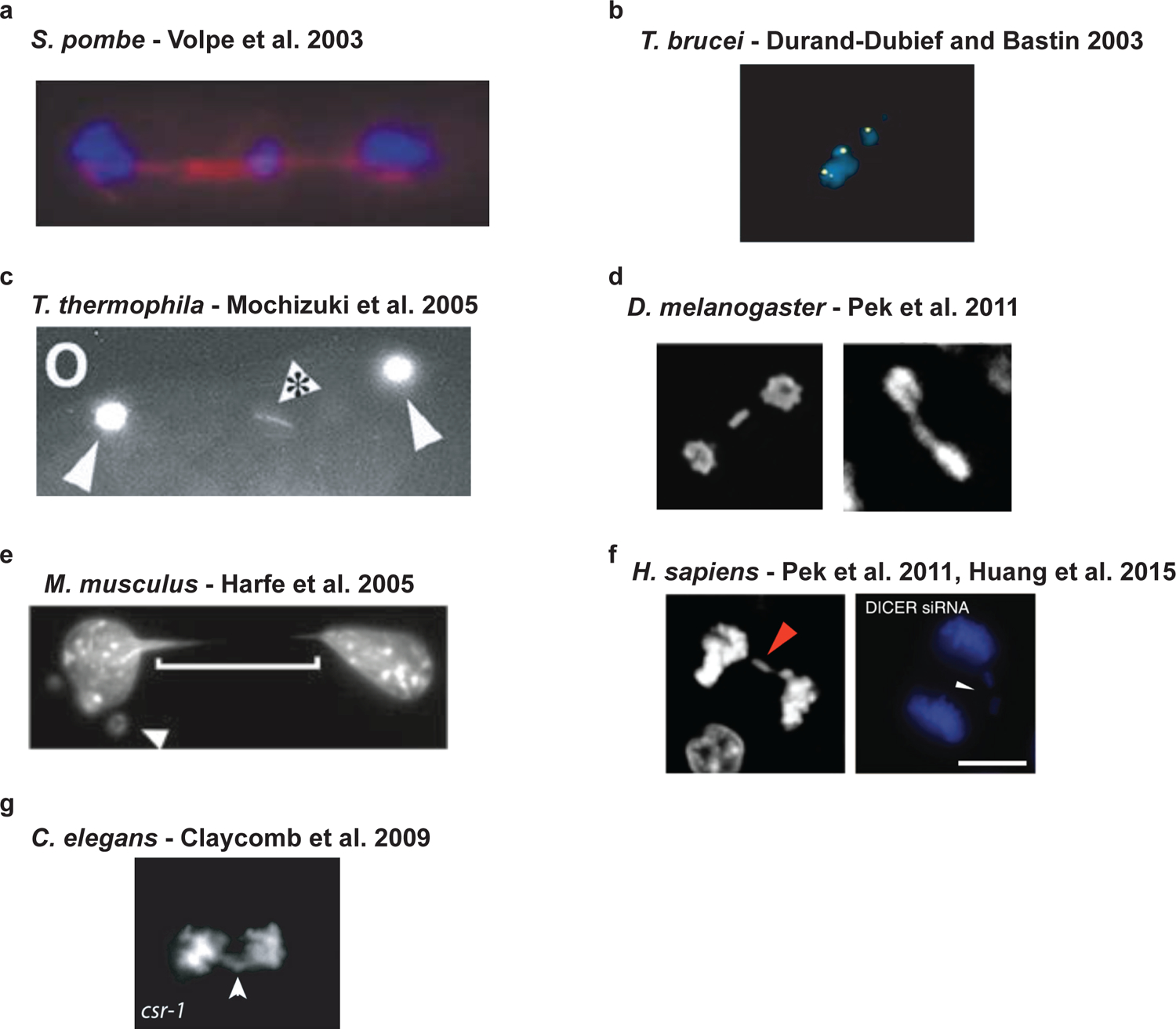
a| Deletion mutants of either the RdRP, Dicer (shown), or Argonaute homologues in S. pombe, result in lagging chromosomes in mitosis and meiosis. b| The deletion of an Argonaute homologue in T. brucei results in lagging chromosomes and segregation defects in mitosis. c| The deletion of a Dicer homologue in T. thermophila results in lagging chromosomes and bridging of genetic material in mitosis and meiosis. d| While the displayed images are of disruptions of belle, the knockout or knockdown of Dicer and Argonaute homologues in D. melanogaster leads to lagging chromosomes in the same cell types - wing disc cells and S2 cell culture. e| The deletion of Dicer1 in mouse fibroblasts results in bridging during mitosis and the formation of micronuclei most likely derived from missegregated chromosomes. f| In human RPE-1 cells, the knockdown of Dicer1 or Ago2 by siRNAs generated lagging chromosomes. In human HeLa cells, the knockdown of Dicer1 also generated lagging chromosomes. g| The knockdown of an Ago homologue (shown) in C. elegans or other factors in the CSR-1 22G-RNA pathway results in lagging chromosomes in the early worm embryo.
