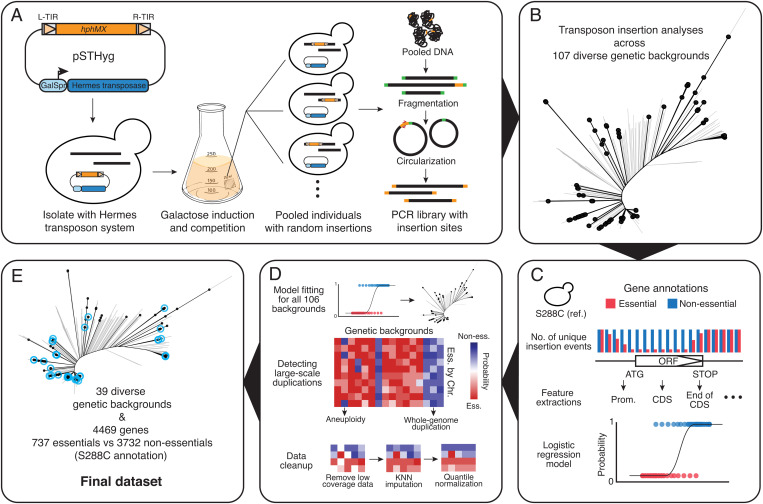
Fig. 1.
Summary of the Hermes transposon saturation procedure. (A) A centromeric plasmid carrying the Hermes transposase and a transposon containing a hygromycin resistance marker (HygMX) are transformed into a haploid isolate background. Random transposon insertions are induced and selected. The mutant pool is then recovered and a PCR library that contains only the insertion sites is constructed and sequenced. (B) Distribution of the selected 107 isolates across the species. The neighbor-joining tree was constructed using biallelic SNPs in the 1,011 yeast collection (26). Selected strains are highlighted in black. (C) A logistic model was constructed using insertion profiles in the reference strain S288C. Gene essentiality annotations were used as a binary classifier, excluding those annotated as involved in galactose metabolism, respiration, and slow growth. (D) The logistic model was applied to insertion patterns in the remaining 106 isolates. Large-scale genome duplications were detected by looking at fitness predictions for all annotated essential genes along each chromosome. Low-coverage regions were removed and then imputed using the k-nearest-neighbor method. The imputed fitness matrix was then quantile-normalized. (E) The final dataset after imputation consists of 39 isolates and 4,469 genes. Strains included in the final dataset are highlighted in blue.