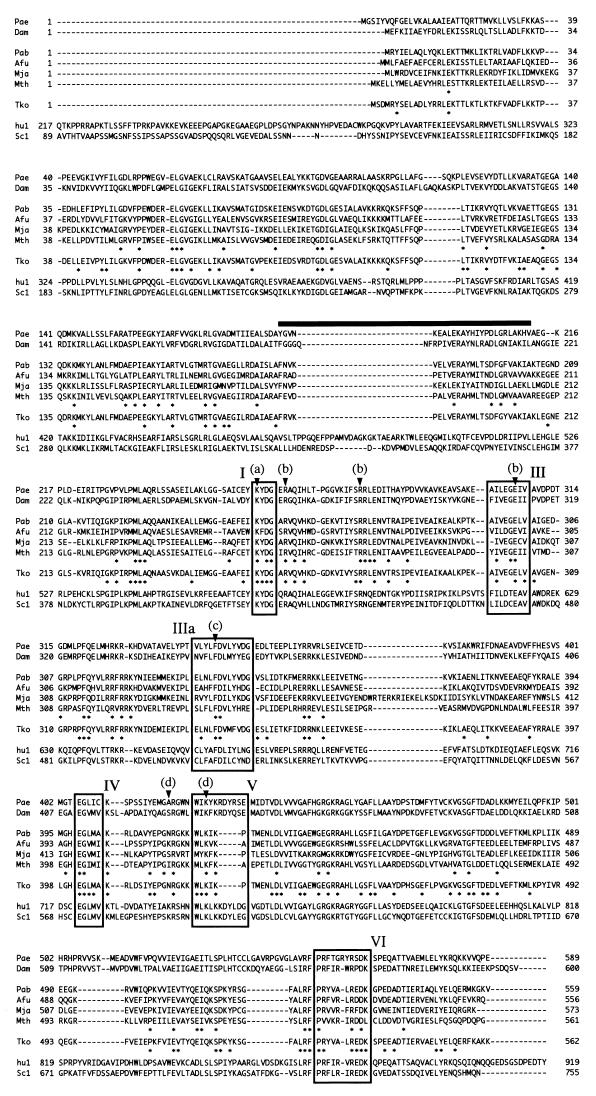
FIG. 2.
Sequence alignment of ATP-dependent DNA ligases from eukaryotes and DNA ligase sequences from archaea. Enzyme, source, and accession number are as follows: Pae, Pyrobaculum aerophilum (U82370); Dam, Desulfurolobus ambivalens (Q02093); Pab, Pyrococcus abyssi (B75173); Afu, Archaeoglobus fulgidus (O29632); Mja, Methanococcus jannaschii (U67474-4); Mth, Methanobacterium thermoautotrophicum (U51624-4); Tko, Thermococcus kodakaraensis KOD1 (this work); hu1, DNA ligase I from humans (NP_000225); and Sc1, DNA ligase I from Saccharomyces cerevisiae (Z74212-1). P. aerophilum and D. ambivalens belong to Crenarchaeota. A. fulgidus, M. jannaschii, M. thermoautotrophicum, and T. kodakaraensis KOD1 belong to Euryarchaeota. Boxes I to VI represent the six motifs commonly found in ATP-dependent DNA ligases mentioned in the text. Arrowheads indicate AMP-binding site (a), ribose binding residues (b), purine ring-stacking residue (c), and phosphate-binding residues (d). The thick bar indicates a region distinct among eukaryotic and archaeal sequences. Asterisks above the Tko sequence indicate conserved residues in archaeal sequences, and those below the Tko sequence indicate conserved residues among Tko, hu1, and Sc1.