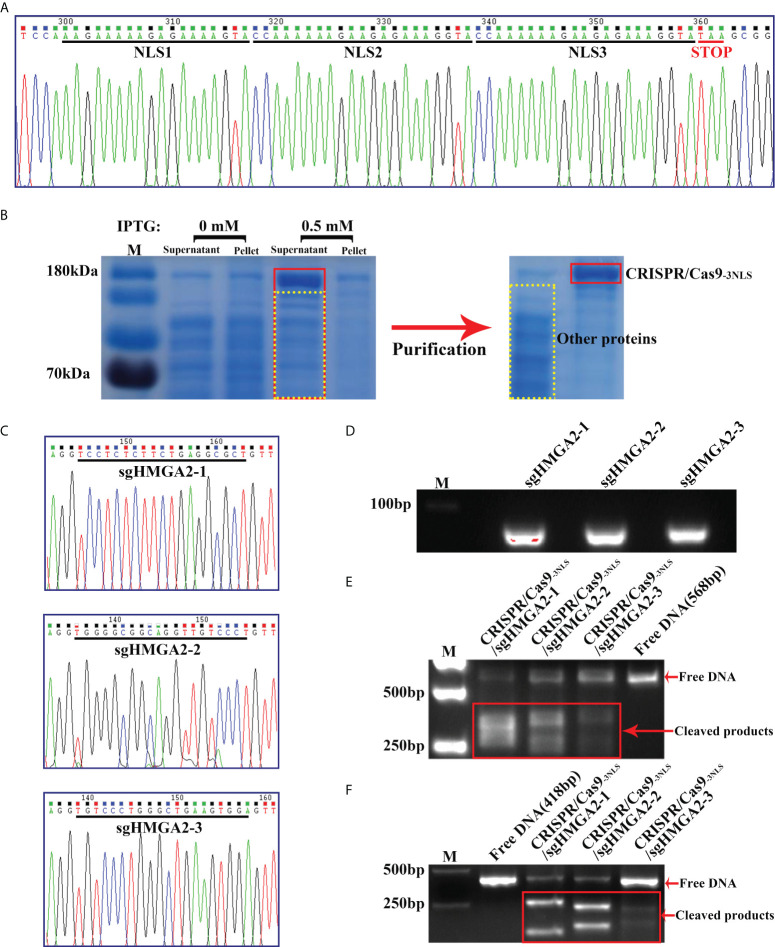
Figure 2.
Preparation of CRISPR/Cas9-3NLS/sgHMGA2 and verification of its cleavage activity (A) The Sanger sequencing result of CRISPR/Cas9-3NLS; black lines indicate the bases encoding two inserted NLSs and one innate NLS, respectively; red line represents the stop code (TAA). (B) SDS-PAGE and Coomassie Brilliant blue staining for analyzing the induced expression and purification of CRISPR/Cas9-3NLS. (C) The Sanger sequencing results of the sgHMGA2 expression plasmid; black lines indicate the bases encoding sgHMGA2-1, sgHMGA2-2, and sgHMGA2-3, respectively. (D) The nucleic acid electrophoresis results of the purified sgHMGA2-1, sgHMGA2-2, and sgHMGA2-3. (E, F) The nucleic acid electrophoresis results of the fragments obtained from cleaving artificially synthesized double-stranded DNA by CRISPR/Cas9-3NLS/sgHMGA2; the length of DNA: 568 bp (E) and 418 bp (F).