Figure 4. Overview of a generalized single-cell workflow: resource investment and key checkpoints.
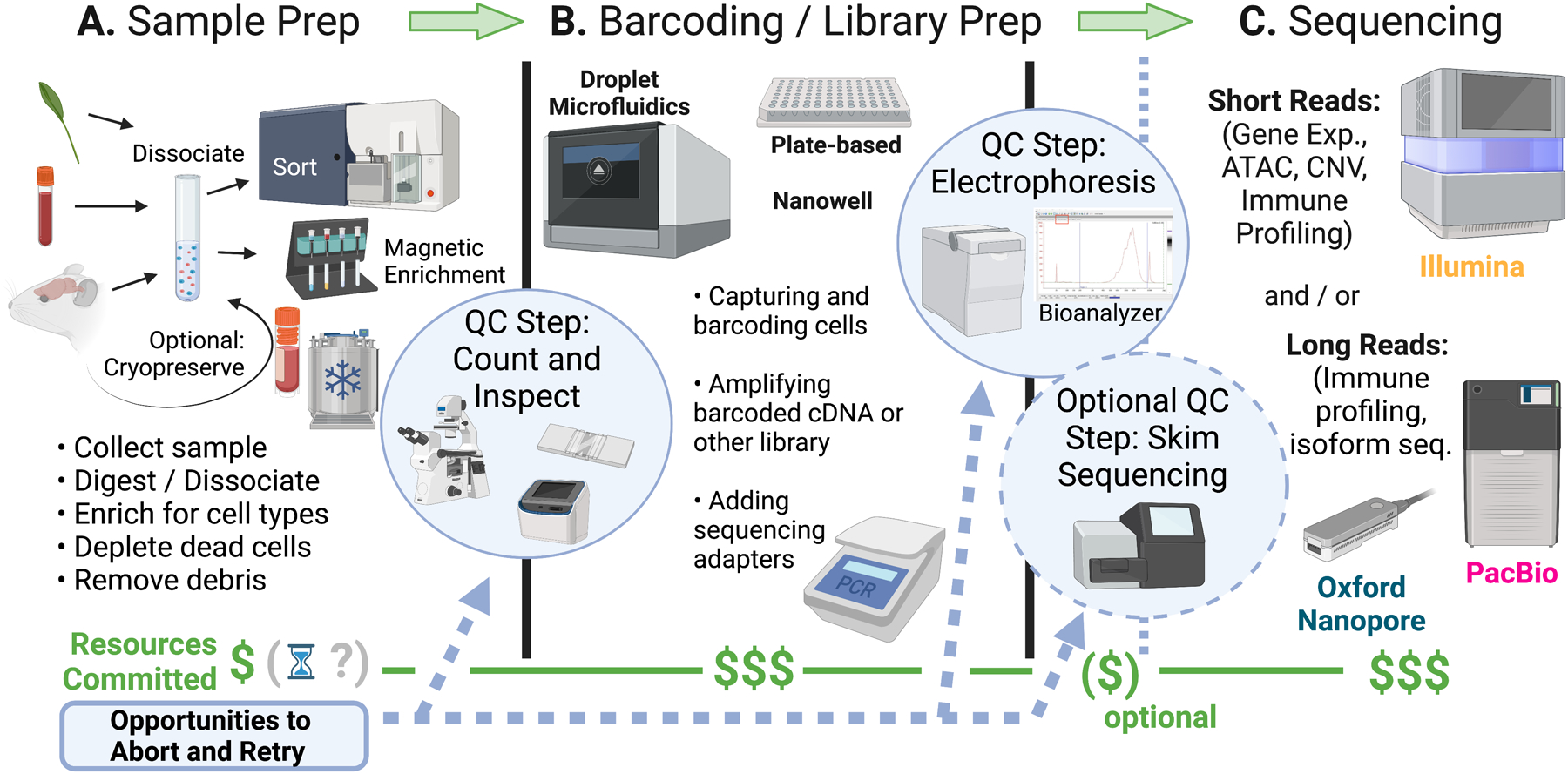
Most single-cell experimental workflows can be subdivided into three distinct phases: sample prep, barcoding and library prep, and sequencing. Each phase entails a significant investment of resources and presents a critical quality control checkpoint that provides opportunities to abort and retry if the samples appear suboptimal. (A) The sample prep stage encompasses all aspects of study design, sample acquisition, storage, and processing upstream of the “genomics” portion of the workflow, and can comprise the majority of the time investment involved in a project. On the day of the experiment, samples are dissociated into single-cell suspensions, and potentially passed through a flow sorter or magnetic column to obtain an enriched population of interest. The cell suspensions should be assessed (e.g., by microscopy) for relevant parameters including purity, viability, cleanliness of the suspension, and clumping. If the suspension looks unsatisfactory or if there are too few intact cells, this is the best time to abort before large amounts of resources are committed in the downstream steps. (B) Barcoding and library prep involves a series of enzymatic reactions that take place in emulsion droplets, PCR plates, nanowells, or other type of isolated compartment. Depending on the library chemistry, this step can consume roughly half of the costs associated with the experiment. Libraries are generally amplified by PCR with the addition of barcoded adapters, and should be assessed at a second QC checkpoint by electrophoresis (e.g., using a Bioanalyzer). (C) Sequencing of the single-cell libraries also consumes a significant percentage of the overall budget. Depending on the application, libraries are sequenced using either short reads (for gene expression, ATAC, CNV, immune profiling, or other applications) on an Illumina instrument, or long reads (e.g., for isoform-resolved RNA-seq, immune profiling) using an Oxford Nanopore or PacBio instrument. For large projects, an optional “skim-sequencing” step can be added for quality control. A few million reads per library is often sufficient to tell whether barcoding proceeded properly, and can provide a crude estimate of captured cell numbers and predicted sample quality.
