Figure 8. Depth requirements.
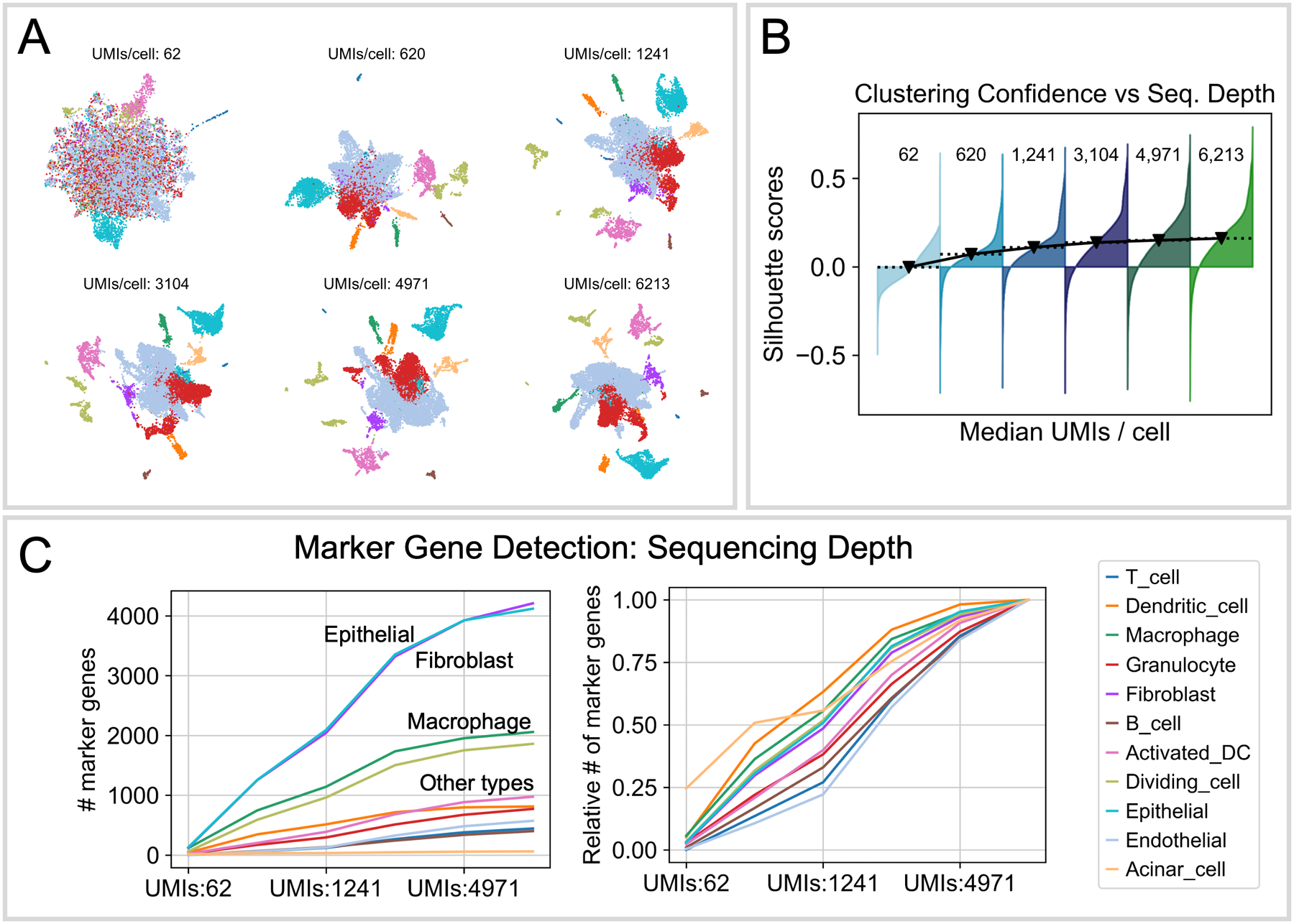
Ideal sequencing depth is dependent upon the project’s goals. For example, clustering and cell type identification tasks in scRNA-seq are relatively robust to shallower sequencing, modeled here by in silico downsampling of a mouse pancreatic tumor dataset (Elyada et al., 2019) to different median numbers of UMIs per cell. Dimensionality reduction by UMAP (A) and clustering confidence (B) demonstrate the impact of sequencing depth on resolving cell types. Distinct clusters (A, colored points) become resolvable with only a few hundred UMIs per cell, with rarer cell types emerging as separate clusters only at higher depths. (B) Unsupervised clustering was run on the downsampled datasets, and inter-cluster silhouette score was calculated as a measure of confidence in cluster assignments. Higher sequencing depth returns only a modest improvement in unsupervised clustering confidence as median UMI counts increase beyond ~2,000/cell. (C) Marker gene detection scales roughly linearly over the full range of subsampled depths. Cell type labels were fixed, and differential expression was performed to detect marker genes across cell types at each subsampled depth. Cell types with high mRNA content, such as epithelial and fibroblast cells, yield more marker genes at low sequencing depth compared with other types (left panel). This phenomenon is typically the result of the fact that a greater proportion of the total UMIs in the dataset come from mRNA-rich cells. Nonetheless, marker gene detection as a function of depth is roughly linear for all cell types, as visualized by normalizing the trend to the maximum number of recovered markers for each type (right panel). This illustrates how sequencing more deeply can help recover more marker genes in cell types with low mRNA content or which comprise only a small proportion of the total library.
