Figure 9. Read length requirements.
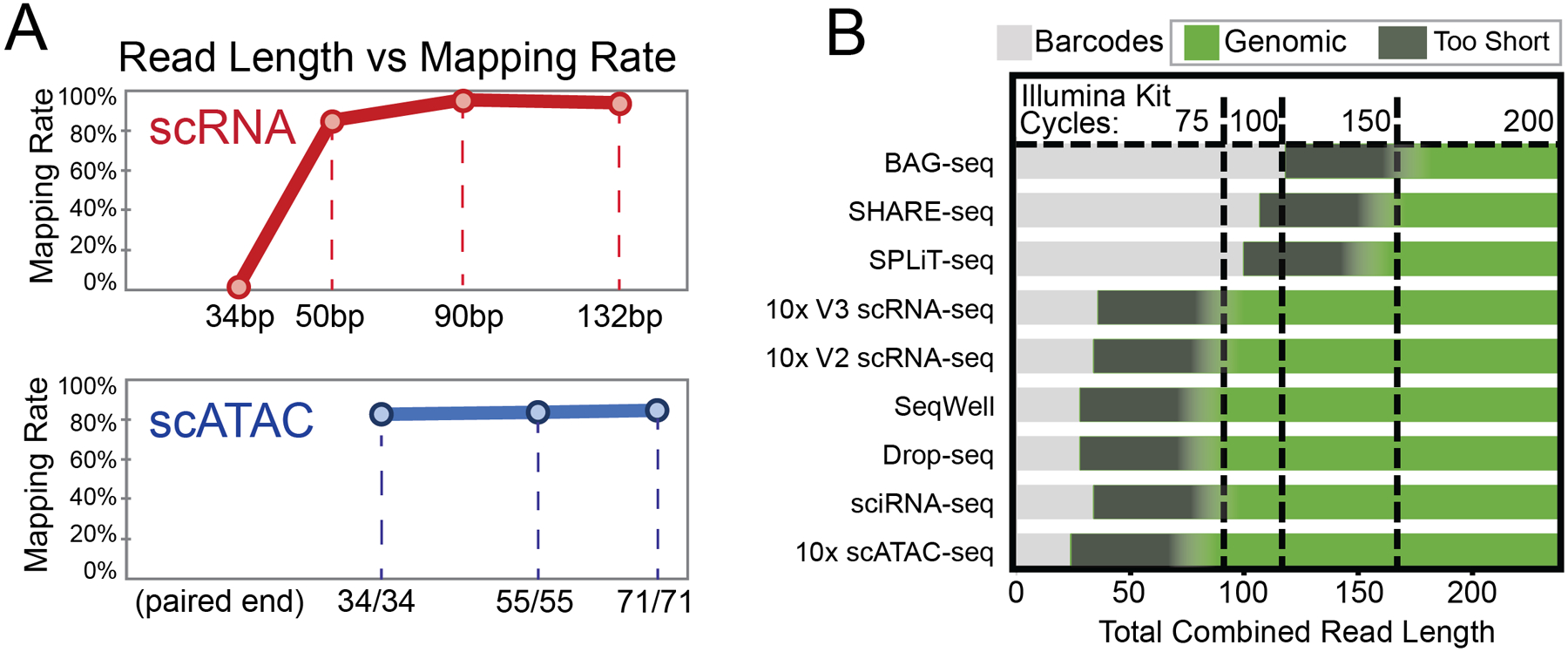
Single-cell modalities require different read lengths for optimal performance. For example, using the 10X Genomics scRNA-seq platform, longer reads return modestly higher mapping rates above a certain minimum threshold length of the gene body read (A, top). The example shown represents human peripheral blood mononuclear cells prepared using 10X Genomics Single Cell 3’ Gene Expression version 3 chemistry and sequenced on an Illumina NextSeq 500 with a 132 bp gene-body read (unpublished data). Reads were trimmed in silico and remapped to assess overall mappability as a function of gene-body read length. Similarly, commonly used read lengths for scATAC-seq are largely indistinguishable in mapping rate (A, bottom). Here, 10X Genomics Single Cell ATAC libraries were created from dissociated mouse pancreatic tumors and sequenced on an Illumina NextSeq 500 with symmetric paired-end reads, informatically trimmed to various lengths (unpublished data). (B) Required read lengths also differ by technology platform as a consequence of their barcode design. Single-cell protocols use a variety of library design strategies, resulting in different required read lengths to cover key library features. For instance, split/pool methods such as BAG-seq, SHARE-seq, and SPLiT-seq generally require 100 or more bases to cover all barcode regions, while droplet methods employ compact barcodes requiring fewer bases, leaving more sequencing reagents available to dedicate to the gene-body or other features in the library amplicon. Combined barcode and minimum mappable genomic read lengths help determine the most efficient kit size to select for sequencing.
