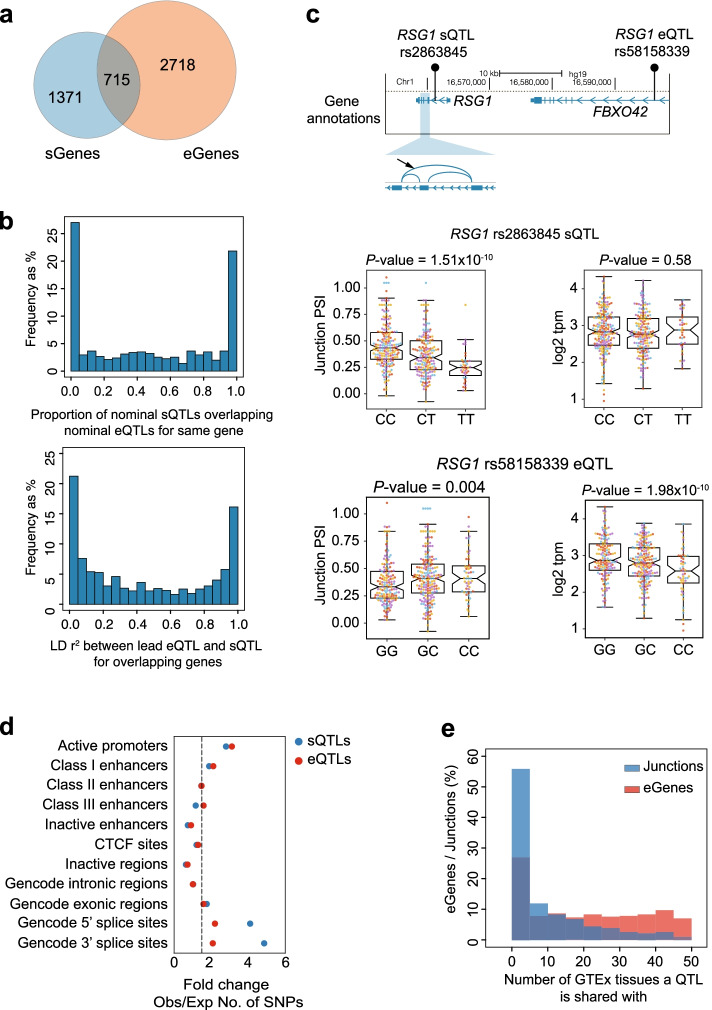
Fig. 2.
sQTLs and eQTLs are distinct genetic signals. a Overlap of sGenes and eGenes. b For 715 genes that have both eQTLs and sQTLs (overlapping genes in a), the top histogram shows the distribution of the percentage of variants shared between sets of nominally significant eQTLs and sQTLs. The bottom histogram shows the distribution of LD (r2) values between the lead eQTL and sQTL. c RSG1 has a distal eQTL, located in an intron of the FBXO42 gene, and an intronic sQTL, both of which are in low LD (r2=0.25). Boxplots represent RSG1 expression and junction PSI values for both sQTL and eQTL, showing that the lead eQTL rs58158339 is not associated with RSG1 splicing and the sQTL rs2863845 is not associated with expression. Boxplots show normalized, batch-corrected expression or junction PSI values stratified by the genotype of the lead QTL (IQR boxes and 1.5 × IQR whiskers). Individual samples are colored according to the human islet dataset they belong to (see color legend in Fig. 1a). Nominal QTL p-values are provided. d Enrichment of sQTL and eQTL variants in different functional genomic annotations using GREGOR. The x-axis represents GREGOR fold change of observed vs. expected number of SNPs at each functional annotation. The dotted line represents 1.5-fold change. e Percentage of eGenes and Junctions with eQTLs or sQTLs at FDR ≤ 1%, respectively, shared in different number of GTEx tissues. We used significant eQTLs and sQTLs from 47 distinct GTEx V8 release tissues