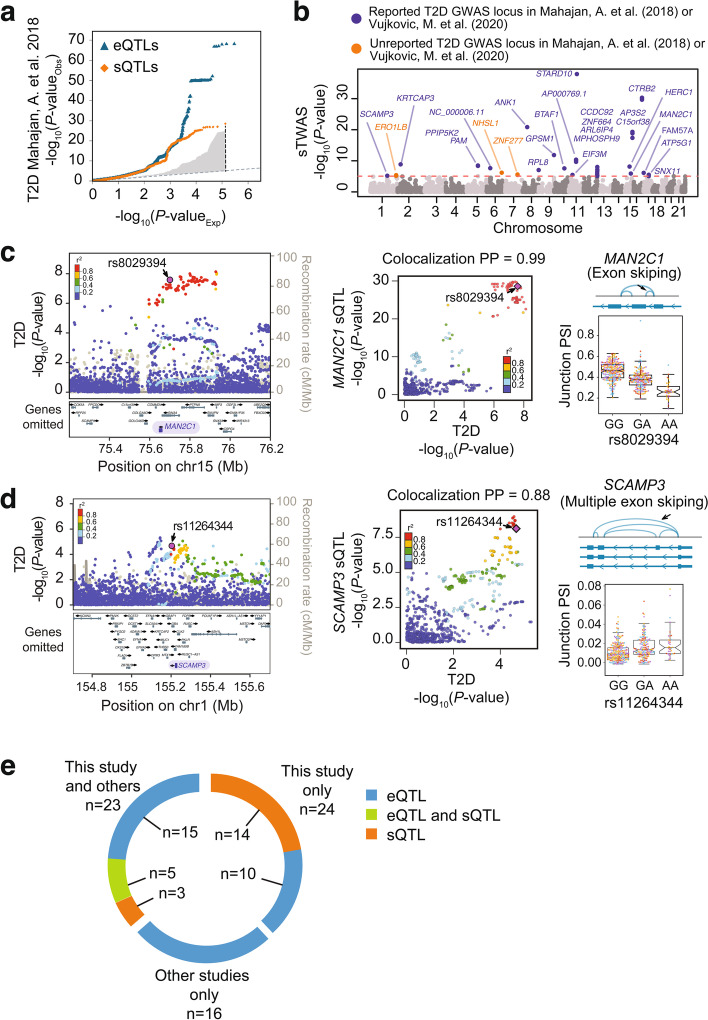
Fig. 3.
Role of human islet splicing variation in T2D susceptibility. a Quantile-quantile (QQ) plot showing observed T2D association p-values in human islet sQTLs (orange dots) and eQTLs (blue dots) against p-values under the null hypothesis. The grey-shaded region represents 1000 p-value distributions (in the -log10 scale) of random sets of control sQTL variants (see the “Methods” section). Each set of control variants matches the number of islet sQTLs plotted. b Manhattan plot of splicing associations with T2D susceptibility (sTWAS). The y-axis shows -log10 TWAS association p-values. Significant sTWAS associations in known T2D GWAS loci are colored in purple, and in previously unreported loci in orange. c, d Regional T2D GWAS signal plots for PTPN9 and PKLR loci, two known T2D susceptibility regions, which showed significant sTWAS signals in MAN2C1 (c) and SCAMP3 (d) genes, respectively. Both splicing QTL effects in MAN2C1 and SCAMP3 are significant at FDR ≤ 1%; see Additional file 3: Table S2. LocusZoom plots show -log10 T2D association p-values and locations in hg19 genome build. LocusCompare scatter plots show sQTL and T2D GWAS association p-values (-log10 scale), illustrating co-localization of variants for both traits. Variants are colored according to the LD correlation (r2) with the lead GWAS variant of the sTWAS association (purple diamond). Boxplots show normalized, batch-corrected junction PSI values on y-axis stratified by the genotype of the lead GWAS variant from the sTWAS association. Boxplots follow the color-to-batch legend from Fig. 1a. e Known T2D loci with target effector transcripts nominated by sQTLs and eQTLs from this study and/or eQTL maps from the InsPIRE consortium