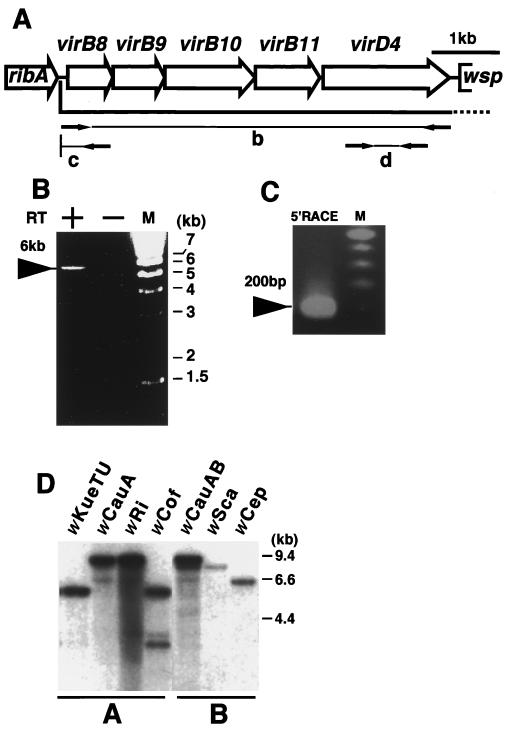
FIG. 2.
Transcriptional analysis of the vir operon and its distribution in Wolbachia strains. M, DNA size marker. (A) Open reading frame map of the wTai vir operon. Open arrows show open reading frames. Arrows in the regions b, c, and d indicate the primers used in the experiments shown in panels B, C, and D, respectively. ribA, GTP cyclohydrolase II; wsp, a surface protein. (B) RT-PCR of the entire region of the vir operon. RT+ and RT− indicate the presence and absence of reverse transcriptase in the reaction mixture, respectively. (C) 5′RACE analysis. (D) Southern hybridization of different Wolbachia strains using virD4 of wTai. wKueTU, wCauA, wRi, and wCof are A group strains, and their insect hosts are E. kuehniella (Tsuchiura strain), the almond moth E. cautella, the fruit fly Drosophila simulans Riverside, and D. simulans Coffs, respectively. wCauAB indicates A and B group strains (wCauA and wCauB, respectively) doubly infecting E. cautella. Two bands were observed in wCauAB, at about 9.4 and 9 kb, while only a single band was observed in wCauA, at 9.4 kb, indicating that wCauB contained virD4 distinct from that of wCauA. wSca and wCep are B group strains infecting the moth Ostrinia scapulalis and Corcyra cepharonica, respectively. All DNA samples were digested with EcoRI.