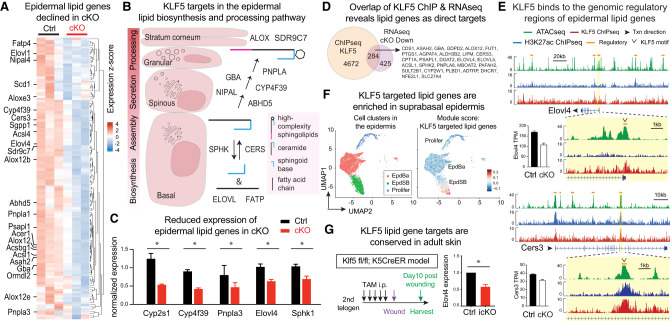
Figure 3.
KLF5 transcriptionally regulates epidermal sphingolipid genes by binding to their genomic regulatory regions. (A) Heat map of differentially expressed genes within the scored lipid metabolic pathways (oxidoreductase, fatty acid biosynthesis, transporter, lipoxygenase, and acyl transferase/hydrolase) highlighting that many genes in epidermal lipid biosynthesis and processing are decreased in cKO compared with Ctrl. (B) Schematic overview of complex sphingolipid biosynthesis and processing in the epidermis. De novo ceramide synthesis occurs in the endoplasmic reticulum of the basal and suprabasal spinous epidermal layers involving ceramide synthase CERS. The inverse process ceramide degradation involves sphingosine kinase SPHK. Preparation of fatty acid substrates involves elongase ELOVL and transporter FATP, among others. Specific to the stratifying epidermis, ceramides are further modified by GBA, NIPAL, PNPLA, AYP4F39, and ABHD5; packaged into lamellar bodies in the granular layer to form high-complexity sphingolipids, including acylceramides; and then secreted into the stratum corneum for postsecretion processing (ALOX and SDR9C7) and ultimately incorporation into the corneocyte lipid envelope and intercellular lipid lamellae. Many more enzymes are involved in the multistep process, and only a few examples are shown here. (C) qPCR validation of epidermal lipid biogenesis and processing genes down-regulated in cKO epidermis compared with Ctrl. n = 3 in each group. Paired t-tests were performed. Data are mean ± SEM. (*) P < 0.05. (D) Venn diagram overlapping KLF5 ChIP-seq peaks (KLF5-bound genes) with Klf5 cKO down-regulated genes as potential KLF5 targets. A list of lipid metabolism genes within the overlap are shown. (E) ATAC-seq (green), H3K27ac ChIP-seq (blue), and KLF5 ChIP-seq (red) tracks showing KLF5-associated putative regulatory elements (denoted as orange lines above the tracks) of selected complex sphingolipid and acylceramide biosynthesis and processing. Bars next to the tracks are RNA-seq transcripts per million reads (TPM) values from Ctrl (black bars) and cKO (white bars) epidermis. (F) scRNA-seq analysis of epidermal populations using the AddModuleScore function of Seurat to denote the high expression of KLF5 targeted sphingolipid metabolism genes (gene list from Fig. 3A; Supplemental Data S2) in suprabasal epidermis. (G) Klf5fl/fl; K5CreER (icKO) and Klf5fl/+; K5CreER (Ctrl) 2-mo-old adult mice were subjected to daily i.p. tamoxifen injection (7.2 uL of 2% tamoxifen per gram of body weight) for 5 d, followed by a 5-mm punch wound (with splint to avoid dermal contraction). Wounded skin regions were harvested 10 d after wounding, and total RNA was extracted for qPCR analysis. N = 3 technical replicates. Paired t-tests were performed. (*) P < 0.05.