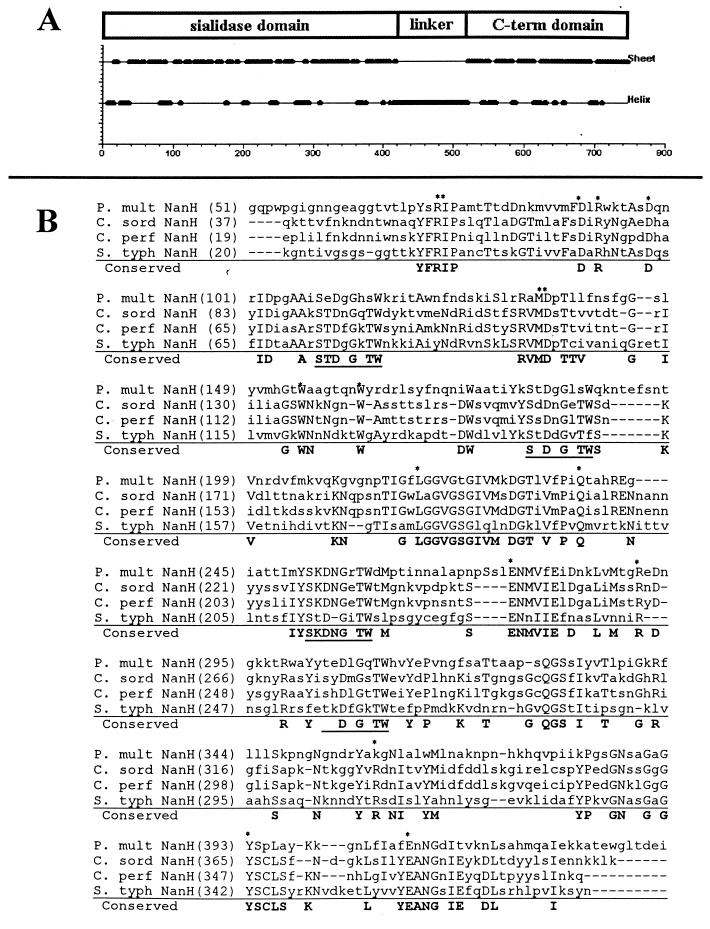
FIG. 1.
Amino acid sequence alignment of P. multocida NanH and related bacterial sialidases. (A) Putative domains of NanH. Protein secondary structure was predicted using the algorithm of Garnier et al. (16). The N-terminal 400 aa of NanH are primarily β sheet, which is consistent with the structure of sialidases. (B) Conservation of specific residues (boldface) between P. multocida (P. mult) NanH and sialidases from Clostridium sordelli (C. sord), C. perfringens (C. perf) (small sialidase), and Salmonella serotype Typhimurium (S. typh). Residues believed to be located in the enzyme active site are marked with asterisks; aspartate boxes are underlined (9, 10). The alignment was generated using the AlignX program of VectorNTI (Informax). The linker and C-terminal domains do not exhibit significant homology to other known proteins.