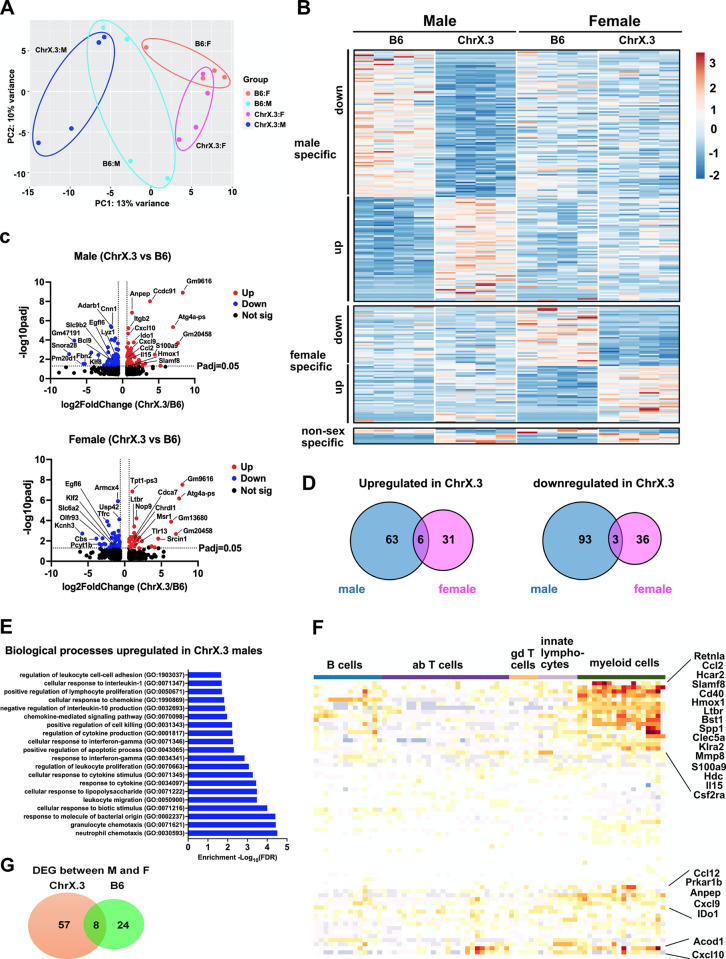
Fig 6. Sex differences in genetically regulated host lung responses in B6 and ChrX.3PWD mice.
Male and female B6 and B6.ChrX.3PWD (ChrX.3) mice were infected with PR8 IAV as in Fig 3. On day 6 post-infection, whole lung RNA was processed for RNA sequencing and analysis as described in Materials and Methods. (A) Principal component analysis was performed in DEseq2, demonstrating clustering of individual samples by sex and genotype, as annotated. (B, C, and D) DEseq2 was used to determine differentially expressed genes between X.3 and B6 mice within males or females, as annotated, using a cutoff of |Log2 (Fold Change)|>0.6 and Padjusted (Padj) <0.05. (B) A row-normalized gene expression heatmap showing DEGs in males, females, or both sexes, as indicated. Volcano plots (C) and Venn diagrams (D) illustrating differential gene expression between ChrX.3 and B6. (E) Biological pathway enrichment analysis on DEGs upregulated in ChrX.3 in males was performed using Gene Ontology as described in Materials and Methods. Top 20 enriched pathways are shown. (F) ImmGen MyGeneSet analysis was performed on DEGs upregulated in ChrX.3 in males as described in the Materials and Methods. Heatmap indicates relative expression of DEGs in cell populations of interest, with each column representing a cell type or cells state within the broader cell categories labeled across the top. (G) A Venn diagram indicating overlap between genes differentially expressed between males vs. females in ChrX.3 and B6 mice.