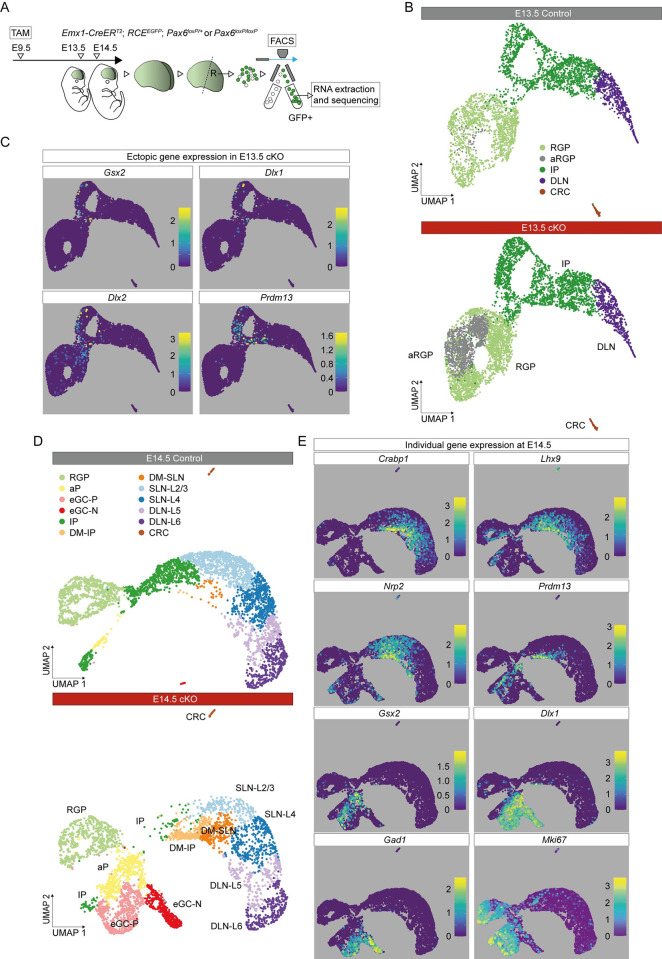
Fig 1. Aberrant cell types and ectopic gene expression in Pax6 cKO cortex.
(A) The experimental procedure: TAM was administered at E9.5; E13.5 and E14.5 rostral (R) cortex was dissociated into single-cell suspension; viable GFP+ cells were selected by FACS for scRNAseq. (B) UMAP plot of the scRNAseq data from Pax6 cKO and control cells at E13.5. Data from the 2 genotypes were analyzed together and then split for visualization. (C) UMAP plots showing log10 normalized expression of selected genes that were ectopically expressed in E13.5 Pax6 cKO cortex. (D) UMAP plots of the scRNAseq data from Pax6 cKO and control cells at E14.5. Data from the 2 genotypes were analyzed together and then split for visualization. (E) UMAP plots showing log10 normalized expression of cell type–selective marker genes at E14.5. aP, atypical progenitor; aRGP, atypical RGP; CRC, Cajal–Retzius cell; DLN, deep layer neuron; DLN-L5 and DLN-L6, layer 5 or layer 6 deep layer neurons; DM-IP and DM-SLN, intermediate progenitor or superficial layer neuron in dorsomedial cortex; eGC-P and eCG-N, proliferating or nonproliferating ectopic GABAergic cells; FACS, fluorescence-activated cell sorting; IP, intermediate progenitor; Pax6 cKO, Pax6 conditional knockout; RGP, radial glial progenitor; scRNAseq, single-cell RNAseq; SLN-L2/3 and SLN-L4, layer 2/3 or layer 4 superficial layer neurons; TAM, tamoxifen; UMAP, uniform manifold approximation and projection.