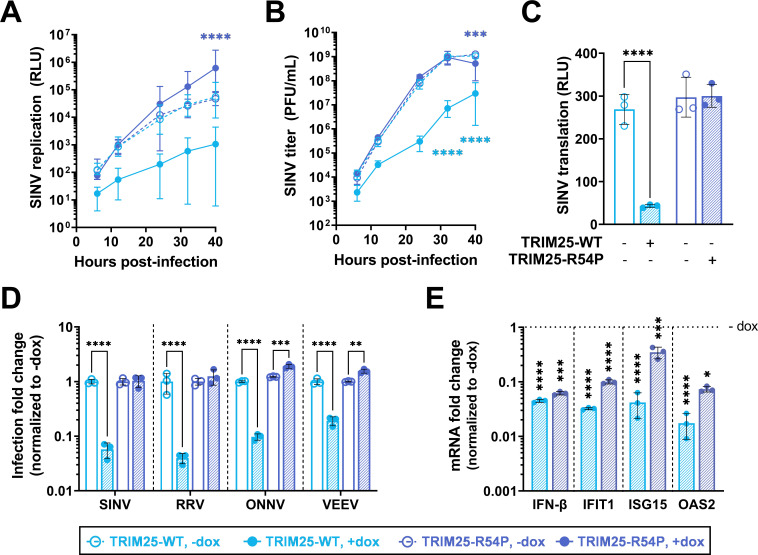
Fig 7. Point mutation in TRIM25 RING domain cripples TRIM25 antiviral activity.
(A-C) Dox inducible TRIM25-WT or -R54P cells were induced for TRIM25-WT or -R54P expression at 1 μg/mL dox. Cells were infected with (A) SINV Toto1101/Luc at an MOI of 0.01 plaque forming unit (PFU)/cell, and lysed at 6, 12, 24, 32, and 40 hours post infection (h.p.i.); data combined from three independent experiments, error bars indicate range; or (B) Sindbis virus (SINV) Toto1101 at an MOI of 0.01 PFU/cell, harvesting supernatant at 6, 12, 24, 32, and 40 h.p.i. for plaque assays; data representative of two independent experiments, error bars indicate range; or (C) SINV Toto1101/Luc:ts6 at an MOI of 1 PFU/cell and lysed at 6 h.p.i. for measurement of luciferase activity; data representative of two independent experiments, error bars indicate standard deviation. (D) Percent infected cells (GFP+) at MOI of 0.01 PFU/cell (SINV 24 h.p.i.; Ross River virus (RRV) 24 h.p.i.; o’nyong-nyong virus (ONNV) 22 h.p.i.; Venezuelan equine encephalitis virus (VEEV) 10 h.p.i.) were normalized to that of the respective cell line without dox (set to one-fold). Asterisks indicate statistically significant differences, calculated using (A-B, D) Two-way ANOVA and Tukey’s multiple comparisons test: **, p<0.01; ***, p<0.001; ****, p<0.0001; (light blue compares WT +/- dox, dark blue compares R54P +/- dox) or (C) Two-way ANOVA and Šidák’s multiple comparisons test: ****, p<0.0001. Data for each virus (demarcated by dashed lines) was statistically analyzed independently. (E) TRIM25 inducible cells were treated with poly(I:C) in the presence or absence of dox, and RNA was harvested for RT-qPCR analysis. mRNA levels of IFN/ISGs in TRIM25-WT or R54P were normalized to that of the respective cell line without dox (set to one-fold, horizontal dotted line). Data representative of two independent experiments. mRNA fold change for each gene (demarcated by vertical dashed lines) was statistically analyzed independently. Asterisks indicate statistically significant differences as compared to the -dox condition (Two-way ANOVA and Šidák’s multiple comparisons test: *, p<0.05; ***, p<0.001; ****, p<0.0001).