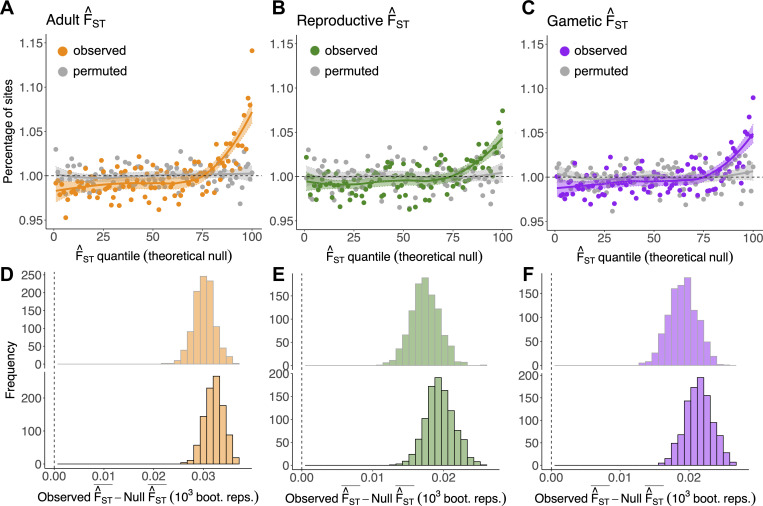
Fig 2.
Polygenic signals of sex-differential selection: Inflation in metrics relative to their nulls. (A–C) Percentage of sites (coloured, observed; grey, permuted) falling into each of 100 quantiles of the theoretical null distributions of adult (A), reproductive (B), and gametic (C). Theoretical null data (x-axes) were generated by simulating values (nSNPs = 1,051,949) from a chi-square distribution with 1 degree of freedom. For each locus, observed and permuted values were scaled by the multiplier of the relevant theoretical null distributions (i.e., the multiplier in Eqs [3A–3C] for adult, reproductive, and gametic , respectively; see Materials and methods). In the absence of sex differences in selection, approximately 1% of observed SNPs should fall into each quantile of the null (dashed line). LOESS curves (±SE) are presented for visual emphasis. (D–F) Difference between the mean of observed and empirical null data for each metric (i.e., adult, reproductive, and gametic , respectively) (top), and the difference between observed and theoretical null data (bottom), across 1,000 bootstrap replicates. Vertical line intersects zero (no difference between observed and null data). As in panels (A–C), values were scaled by the relevant theoretical null distributions. The code and data needed to generate this figure can be found at https://github.com/filipluca/polygenic_SA_selection_in_the_UK_biobank and https://zenodo.org/record/6824671. SNP, single-nucleotide polymorphism.