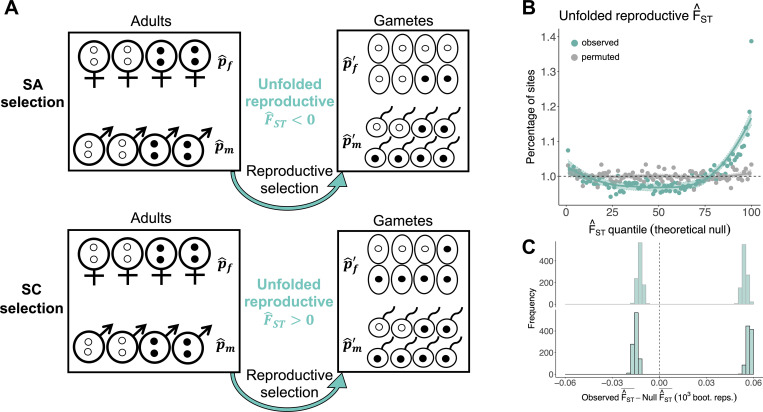
Fig 3. Partitioning signals of sex-differential selection into SA and SC components reveals their joint contributions.
(A) As in Fig 1, , and depict sex-specific frequency estimates for a given allele at different stages of the life cycle. Under SA selection (top), the white allele is female-beneficial and the black allele is male-beneficial, which tends to generate negative values of unfolded reproductive . Under SC selection (bottom), the black allele is beneficial in both sexes, which tends to generate positive values of unfolded reproductive . (B) Percentage of sites (turquoise: observed; grey: permuted) falling into each of 100 quantiles of the theoretical null distributions of unfolded reproductive . Theoretical null data (x-axes) were generated by simulating values (nSNPs = 1,051,949) from the null (i.e., the product of 2 standard normal distributions). In the absence of sex-differential selection, approximately 1% of observed SNPs should fall into each quantile of the null (dashed line). LOESS curves (±SE) are presented for visual emphasis. (C) Difference, for unfolded reproductive , between the mean observed and empirical null data (top) and between observed and theoretical null data (bottom), across 1,000 bootstrap replicates. The vertical line intersects zero, indicating no difference between the observed and null data. Differences between observed and null data were obtained separately for negative and positive values of unfolded reproductive . This illustrates that there is enrichment of SNPs in both tails of the null. The code and data needed to generate this figure can be found at https://github.com/filipluca/polygenic_SA_selection_in_the_UK_biobank and https://zenodo.org/record/6824671. SA, sexually antagonistic; SC, sexually concordant; SNP, single-nucleotide polymorphism.