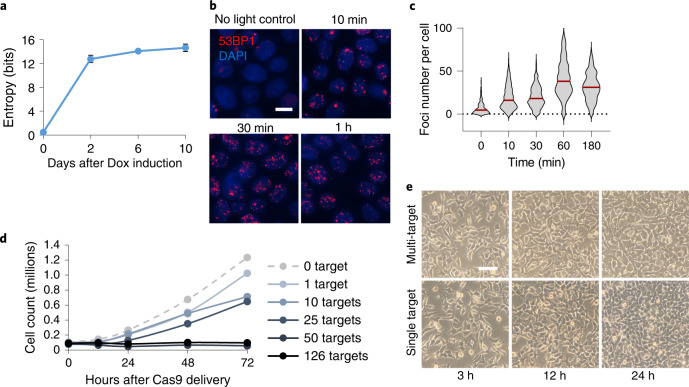
Fig. 7. Quantification of entropy and DNA damage generated by mgRNAs.
a, Cumulative Shannon entropy for the ten-target mgRNAs across the eight on-target sites with detectable indels. Higher entropy corresponds to higher mutation diversity that is necessary for effective cellular barcoding. n = 3 biologically independent experiments, data presented as mean ± s.d. b, Representative images of 53BP1 immunofluorescence microscopy as a function of time after ‘GG’ multi-target Cas9 activation in HEK293T cells using the light-inducible vfCRISPR system. Cas9/gRNA was electroporated into cells 12 h before light-based Cas9 activation. Scale bar, 10 μm. n > 300 cells examined over three independent experiments. c, Quantification of number of 53BP1 foci in each cell from b at all evaluated timepoints. d, Cell counts over time (12 h, 24 h, 48 h and 72 h) after delivery of Cas9 into HEK293T cells with mgRNA targeting various numbers of expected genome-wide sites (0, 1, 10, 25, 50 and 126 expected sites). Data points are the average of two biologically independent samples. e, Representative microscopy images of HEK293T cells at different timepoints (3 h, 12 h and 24 h) after electroporation of Cas9 with multi-target (‘GG’) versus single-target (targeting ACTB) gRNA. Scale bar, 50 μm. Source numerical data are available in source data.