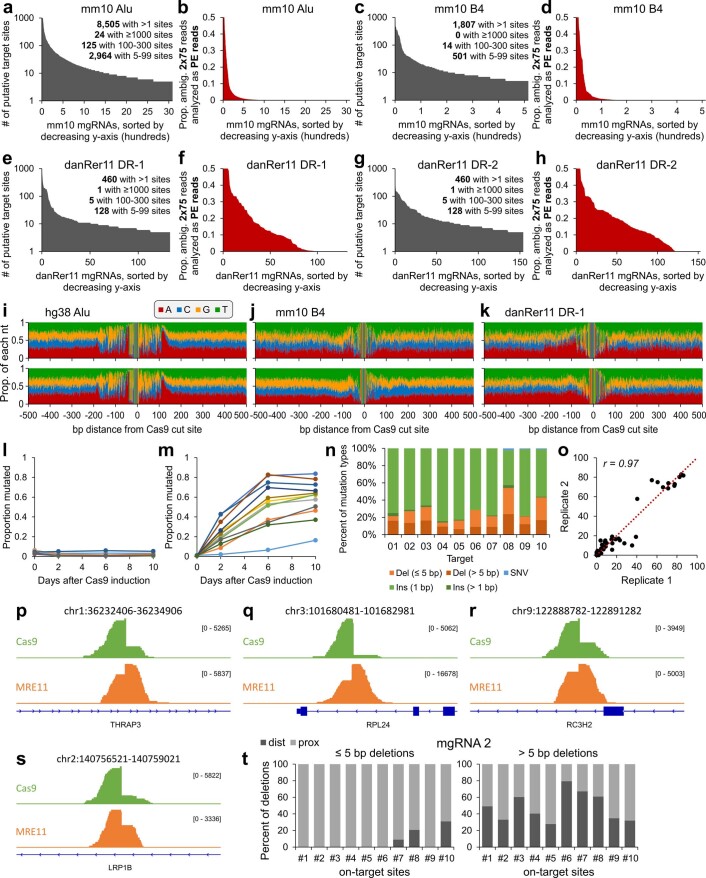
Extended Data Fig. 1. Additional in silico and experimental characterization of multi-target gRNAs.
a,b, Same as Fig. 1b and Fig. 1h, respectively, but for the Alu SINE in the mouse mm10 genome. c,d, Same as Fig. 1b and Fig. 1h, respectively, but for the B4 SINE in the mouse mm10 genome. e,f, Same as Fig. 1b and Fig. 1h, respectively, but for the DR-1 SINE in the zebrafish danRer11 genome. g,h, Same as Fig. 1b and Fig. 1h, respectively, but for the DR-2 SINE in the zebrafish danRer11 genome. i-k, Same as Fig. 1k, but for two different mgRNAs from the (i) Alu SINE in the human hg38 genome, (j) B4 SINE in the mouse mm10 genome, and (k) DR-1 SINE in the zebrafish danRer11 genome. l, No-dox control mutation curve. Dox-inducible Cas9 cells genomically integrated with a 10-target mgRNA (same cells as in Fig. 1l) were grown and passaged without dox exposure. Cells were harvested at different time points (0, 2, 6 and 10 days). m, Mutation curve of 20-target mgRNA in HeLa cells with dox-inducible Cas9. Figure details are the same as Fig. 1l. n, Mutation signatures for 10-day samples from m. Figure details are the same as in Fig. 1m. o, Same reproducibility analysis and figure details as in Fig. 1n but using the mgRNA from m. p-s, For all 4 target sites, the PAM-distal side is oriented on the left. (p-r) The first three examples exhibit Cas9 PAM-distal bias and MRE11 PAM-proximal bias. However, the last example (s) in chr2:140756521-140759021 illustrates an exception where both Cas9 and MRE11 exhibit PAM-distal bias. t, Same as Fig. 3j, but with the 20-target mgRNA 2. Source numerical data are available in source data.