Fig. 6. TERT promoter duplications mimic hotspot mutations for GABP tetramer recruitment.
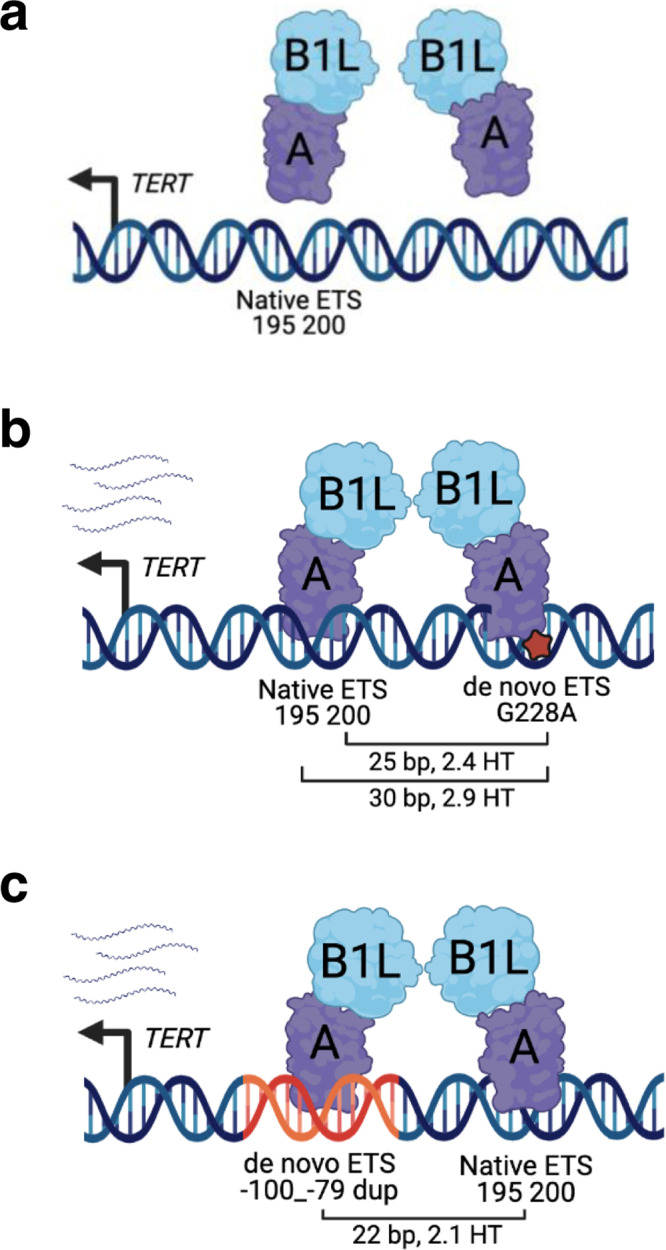
a Schematic of the wildtype TERTp. The native ETS motif is shown relative to the TERT ATG translational start site. GABPB1L-GABPA heterodimer is not bound to the native ETS motif and an inactive TERTp. b Schematic of the TERTp with a hotspot mutation. The native ETS motif and de novo ETS motifs (hotspot mutation) are shown relative to the TERT ATG. Distance in base pair (bp) between the native ETS motif and de novo motifs (hotspot mutation) is shown along with the associated helical turns (HT). GABPB1L-GABPA tetramer binding to the native ETS motif and de novo ETS motif (G228A, hotspot mutation) to activate the TERTp. c Schematic of the TERTp duplication. Native ETS motif and de novo ETS motif (c.−100_−79, duplication) are shown relative to the TERT ATG. Distance (bp) between the native ETS motif and de novo ETS motif (duplication) is shown along with the associated helical turns (HT). GABPB1L-GABPA tetramer binding to the native ETS motif and de novo ETS motif (c.−100_−79, duplication) to activate the TERTp. The figures were created with BioRender (a–c).
