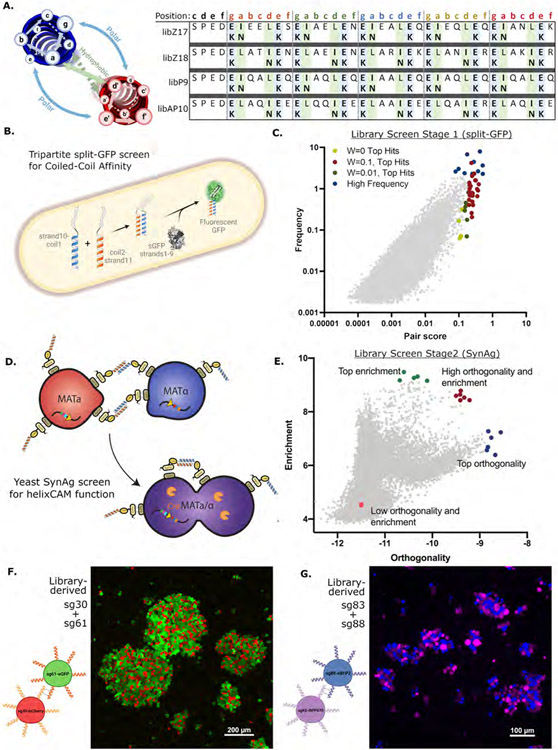
Figure 3. Rational Design of helixCAM-optimized Coiled-Coil library and two-stage screen for helixCAM performance.
A. Table of amino acid substitutions for each of four template-derived helixCAM libraries. Each template consists of five heptads, within which either the “g”, “a” and “e” position or the “g”, “d”, and “e” positions are varied to form new electrostatic and hydrophobic interactions. B. Design of the tripartite split-GFP assay for CC affinity. One CC library was fused to β-strand 10 and another to β-strand 11 of the tripartite split-GFP. Interaction between CCs stabilizes a fluorescent GFP protein. C. Graph of CC candidate pairs’ frequency (in percent) in the population versus their pair score (graph uses W=0.1). The pair score represents a pair’s specificity to each other and is adjusted with a weight parameter W (Detailed in methods). The top 30 hits using three different W’s, along with several high-frequency pairs (totaling 102 individual CCs) were selected for subsequent screening. D. Design of modified yeast SynAg mating assay for helixCAM-compatible CC selection. Haploid yeast cells MATa or MATα expressing surface-presented CC candidates were mixed. CC binding induces haploid cells to mate, creating a diploid cell that survives dual auxotrophic selection. The fusion of cells also leads to the expression of the Cre recombinase (orange), which integrates the two CC constructs and their barcodes into the same DNA strand. E. Stage 2 CC candidate pairs’ enrichment versus their orthogonality. Enrichment is the log of the observed frequency of the pair as a ratio to their individual frequency in the pre-mated populations, whereas orthogonality is the log of the frequency of the pair divided by the total observations of each of the two CCs in the pair. Two pairs, sg30/sg61 and sg83/sg88, were selected from the red group for helixCAM use. F. Large aggregates of sg30mCherry and sg61eGFP K562 cells. Image was taken at 20X magnification with a 3x3 tile and subsequently cropped. G. Aggregates of sg83iRFP670 and sg88eBFP2 K562 cells. Image was taken at 20X magnification and presented without cropping.