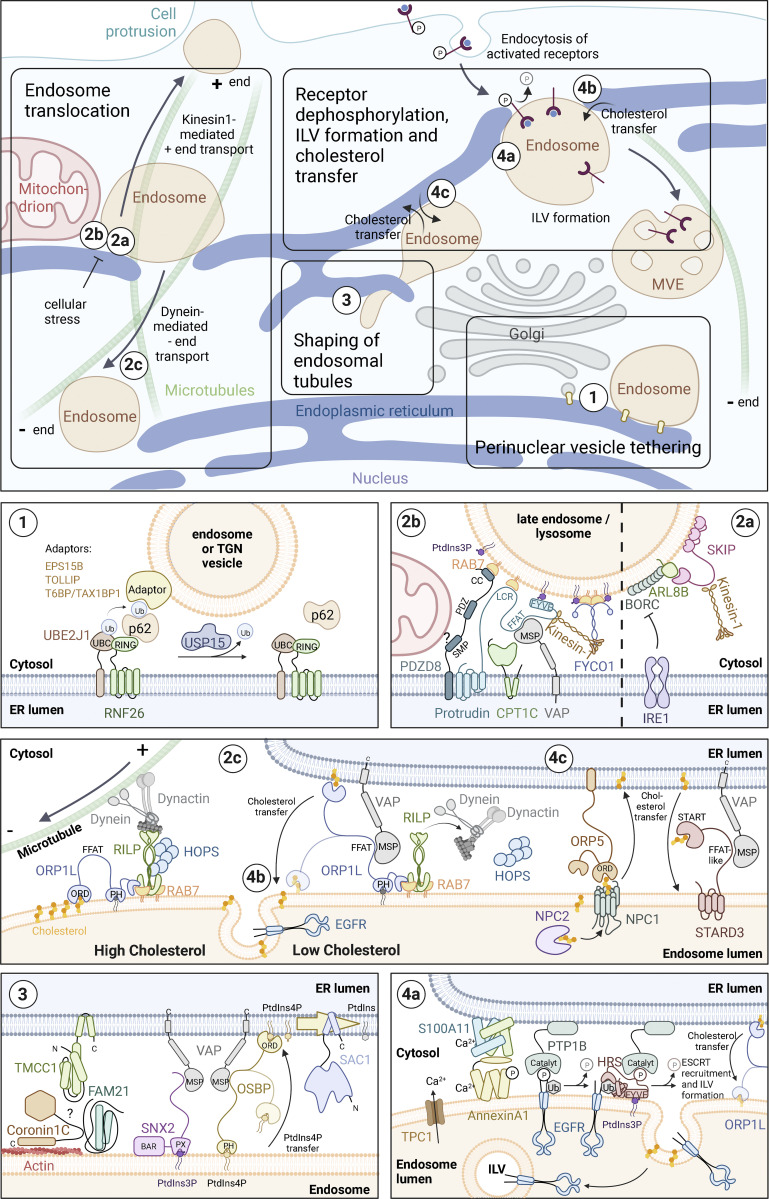
Figure 1.
ER-mediated control of endosome dynamics. Overview of cell biological functions of ER-endosome contact sites and the involved molecules. The molecular composition of ER–endosome contact sites. OSBP, ORPs, and VAPs function as dimers or multimers. For simplicity, this is not displayed in the figure. VAP family members (see text box) are depicted as “VAP.” (1) Perinuclear vesicle tethering: The E2 ubiquitin-conjugating enzyme UBE2J1 activates the E3 ubiquitin ligase RNF26, which then ubiquitinates SQSTM1/p62. Ubiquitinated SQSTM1/p62 in turn binds to organelle-specific adaptor proteins, such as T6BP/TAX1BP1, on TGN vesicles and EPS15B or TOLLIP on endosomes. The release of the tethered vesicles is mediated by the deubiquitinase USP15. (2) Endosome translocation. (2a) The BORC complex recruits the small GTPase ARL8B to endosomes, which in turn recruits and activates the Kinesin-1 adaptor protein SKIP/PLEKHM2, resulting in plus-end directed movement of endosomes and lysosomes. Upon ER stress, IRE1 inhibits BORC-dependent anterograde endosome translocation. (2b) The ER-resident protein Protrudin contacts endosomes by binding to RAB7 and PtdIns3P. At these contact sites, Protrudin mediates the hand-over of Kinesin-1 to the endosomal adaptor protein FYCO1, allowing plus-end translocation of endosomes along microtubules. The activity of Protrudin can be regulated by CPT1C, which promotes anterograde endosome transport under nutrient-rich conditions and blocks it under cellular stress conditions. PDZD8 interacts with Protrudin and RAB7, also mediating ER-endosome contact. In addition, PDZD8 might mediate contact with mitochondria. (2c) Endosomes containing high levels of cholesterol move along microtubules in the minus-end direction by dynein/dynactin motor proteins, which connect to the endosome through RILP, RAB7, and ORP1L. Under low concentrations of cholesterol, ORP1L makes contact with VAP in the ER, which leads to the dissociation of dynein/dynactin and the HOPS complex. ER-endosome contact enables ORP1L to transfer cholesterol from the ER to endosomes. Sufficient levels of endosomal cholesterol are a prerequisite for ILV formation (see also legend to 4b). (3) Shaping of endosomal tubules: The formation of recycling tubules requires transient accumulation of PtdIns4P on endosomes to allow WASH-dependent actin nucleation and retromer function. OSBP interacts with PtdIns4P on endosomes via its PH domain and tethers endosomes to the ER via interaction with VAP. PtdIns4P is then dephosphorylated by the ER-resident lipid phosphatase SAC1, securing a transient PtdIns4P pool on endosomes. WASH is linked to the retromer by its subunit FAM21, which marks the site of tubule scission. The PtdIns3P-binding retromer subunit SNX2 is also able to interact with the ER through VAP. The ER protein TMCC1 and Coronin 1C on endosomes are required for contact site formation and fission of WASH-containing endosome tubules. It is not known whether Coronin 1C and TMCC1 interact directly, or if there are additional proteins required to generate these membrane contact sites. (4) Receptor dephosphorylation, ILV formation, and cholesterol transfer. (4a) EGFR-induced phosphorylation of Annexin A1 induces the formation of Annexin A1/S100A11-mediated ER-endosome contact sites, aided by the local increase in Ca2+ through the endosomal Ca2+ channel TPC1. PTP1B in the ER dephosphorylates EGFRs and ESCRT-0, facilitating the sorting of EGFRs into forming ILVs. (4b) In addition, Annexin A1/S100A11-mediated ER-endosome contact sites facilitate cholesterol transfer from ER to forming ILVs by ORP1L (see also legend to 2c). (4c) STARD3 and its paralog STARD3NL (not shown) mediate cholesterol transfer from ER to EGFR-negative endosomes. ORP5 facilitates cholesterol transport from endosomal membranes to the ER. The cholesterol is provided by NPC2 and NPC1, which interacts with ORP5, forming an ER–endosome contact. Direct shuttling of sterols using the ORD domain of ORP5 remains to be confirmed (Santos et al., 2020).