Figure 1.
Generation of functional human macrophages from pluripotent stem cells
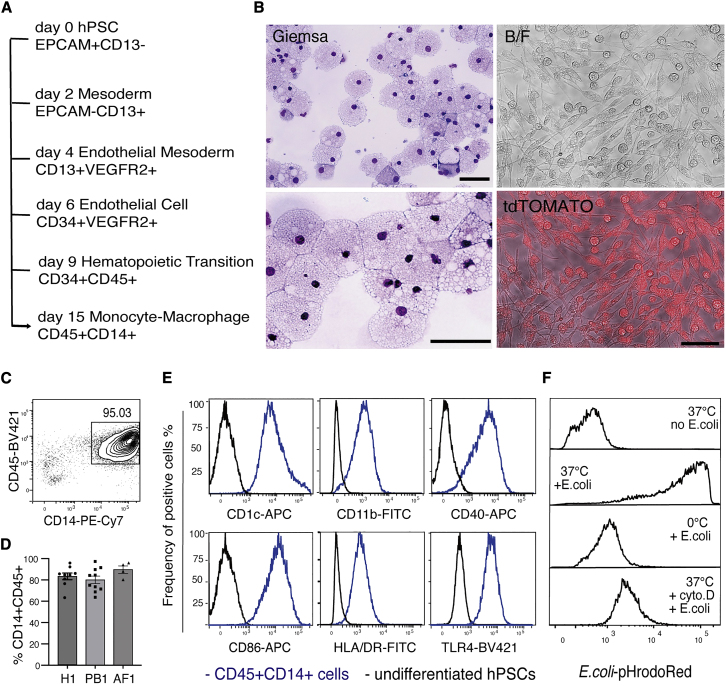
(A) Roadmap of hPSC differentiation to functional macrophages in vitro.
(B) Morphological analysis for hPSC-derived macrophages. Panels show cyto-spin preparations of May-Grunwald-Giemsa-stained macrophages (left) and a bright field (BF) and a BF fluorescence-merged (tdTOMATO) image of macrophages generated from an hPSC line that constitutively expresses a tdTOMATO transgene. Scale bar, 50 μm.
(C) Flow cytometric analysis showing the expression of CD45 and CD14 on cells from differentiation cultures at day 15. The frequency of CD45+CD14+ cells is indicated.
(D) Histograms summarizing the frequency of CD45+CD14+ cells obtained from three independent hPSC lines over more than four independent experiments: H1, n = 9; PB1, n = 10; AF1, n = 4. Data shown as mean ± SEM, non-significant, examined by one-way ANOVA test.
(E) Flow cytometry analysis of CD45+CD14+ cells (blue line) for the expression of surface markers typically associated with functional macrophages. Undifferentiated hPSCs were used as negative controls (black line).
(F) Flow cytometry analysis of hPSC-derived macrophages incubated with pHrodoRed-conjugated E. coli bioparticles under the conditions indicated. Incubation of E. coli bioparticles with hPSC-macrophages was conducted at 37°C (37°C + E. coli). Control groups: hPSC-derived macrophages at 37°C (37°C no E. coli), incubation of E. coli bioparticles with hPSC-derived macrophages at 0°C (0°C + E. coli), incubation of E. coli bioparticles with macrophages with cytochalasin D treatment at 37°C (37°C + cyto. D + E. coli).