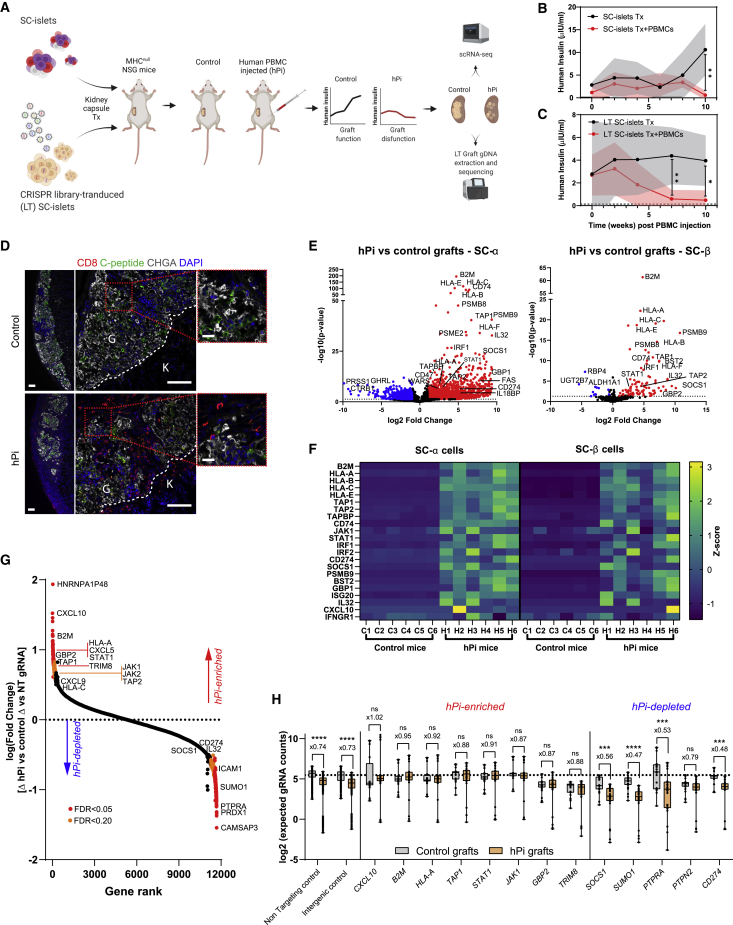
Figure 1.
Single-cell transcriptional profile and whole-genome CRISPR screen of SC-islet grafts in an in vivo humanized model
(A) SC-islets or CRISPR library transduced (LT) SC-islets were transplanted in MHCnull NSG mice. Half of each mice cohort was injected with human PBMCs, and human insulin was monitored until graft failure was observed. Grafted cells were then extracted (week 10 post PBMCs) and analyzed by scRNA-seq for gene expression, or by gDNA sequencing for gRNA abundance.
(B and C) SC-islet graft failure was assayed in fasted mouse blood by human insulin detection over time, 30 min post glucose.
(B) n = 6–8 per group of SC-islet transplanted mice.
(C) n = 6 per group of LT SC-islet transplanted mice.
(D) Immunofluorescence (IF) staining of kidney SC-islet grafts sections at week 10 after PBMC injection. Bars represent 100 μm in left (×5) and center (×20) and 20 μm in magnified view (right). Kidney (K) and graft (G) margins are outlined. CHGA, chromogranin A.
(E and F) scRNA-seq analysis of SC-islet grafts.
(E) Volcano plot of differential expressed genes in SC-β and SC-α in hPi versus control grafts.
(F) Differential expression of selected genes in different populations, presented as a heatmap. Each row specifies a Z score of the specified gene in all graft samples, in the indicated endocrine population.
(G) Analysis of enriched and depleted gene KOs. Rank is plotted against fold changes (hPi versus control) of gRNA counts (×4 integrated per gene) relative to integrated non-targeting (NT) gRNA counts (×941). Significant genes are color coded based on false discovery rate (FDR) as indicated.
(H) Boxplot presenting individual gRNAs counts (full model predictions) from mice replicates (n = 6 per condition times n = 4 targeting gRNAs, n = 85 for NT gRNAs, or n = 50 for intergenic gRNAs) with genes of interest with positive and negative enrichment in screen. Box lines represent median values. Dashed line represents mean of NT gRNA counts in control mice. Error bars or shaded areas are mean ± SD; ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001, unpaired two-tailed t test.