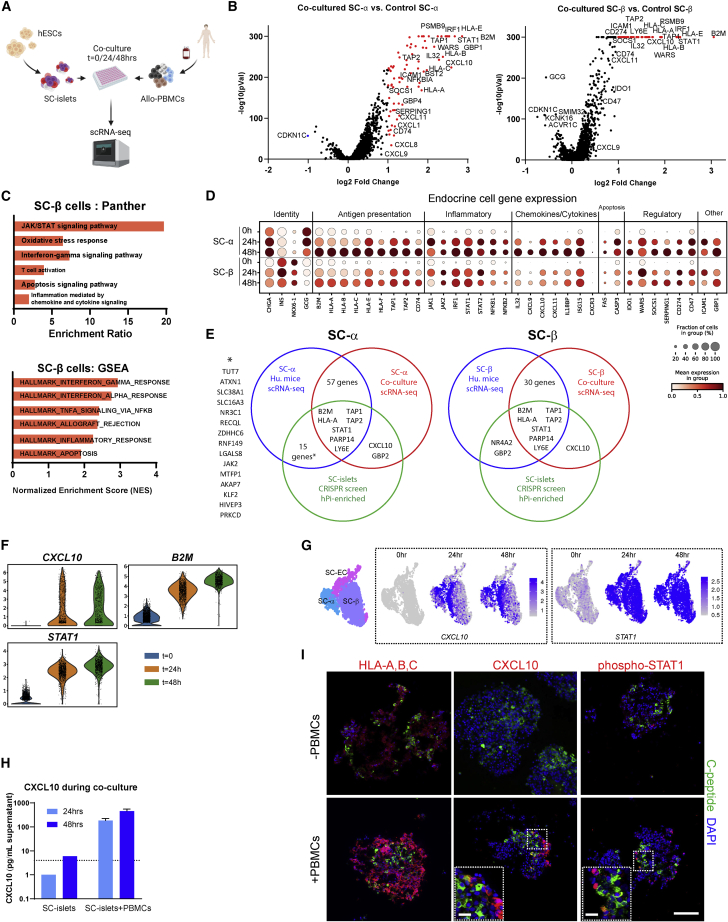
Figure 2.
Early response of immune-challenged SC-islets profiled by single-cell transcription analysis after co-culture with human allogeneic PBMCs
(A) hESC-derived SC-islets were co-cultured with human allogeneic PBMCs (n = 2 donors) for 0, 24, and 48h, followed by scRNA-seq for gene expression.
(B) Volcano plot of differential expressed genes in SC-α or SC-β after 24-h co-culture with PBMCs compared with control (t = 0).
(C) Pathway analysis and gene set enrichment analysis (GSEA) of upregulated genes in co-cultured SC-β (48 h).
(D) Dot plot representing expression of selected inflammatory genes in groups of SC-α and SC-β over time in co-culture with PBMCs.
(E) Venn diagrams feature significantly upregulated genes (log2 fold change >1 and adjusted p values <0.05) obtained from in vivo (blue) and in vitro (red) SC-α/SC-β scRNA-seq data (Figures 1 and 2) that are common to CRISPR screen hits (positively enriched in hPi-mice, log2 fold change >1) (green).
(F) Violin plots of SC-β timed expression of selected genes. See also Figure S2F.
(G) UMAP plots of SC-islet cells expressing CXCL10 or STAT1 over time in co-culture with PBMCs. Specific endocrine cell type clustering is indicated.
(H) ELISA for human CXCL10, from supernatant of co-culture of SC-islets and PBMCs. n = 2 donors. Error bars are mean ± SD. Dashed line is the lower detection limit, while any data below it is extrapolated.
(I) IF staining of SC-islet clusters ±48-h co-culture with PBMC. C-peptide staining (green) for SC-β and DAPI (blue) for nuclei. Bars represent 100 μm in main panels and 50 μm in magnified panels.