Figure 4.
Generation and performance of CXCL10 KO and STAT1 KO hESC lines
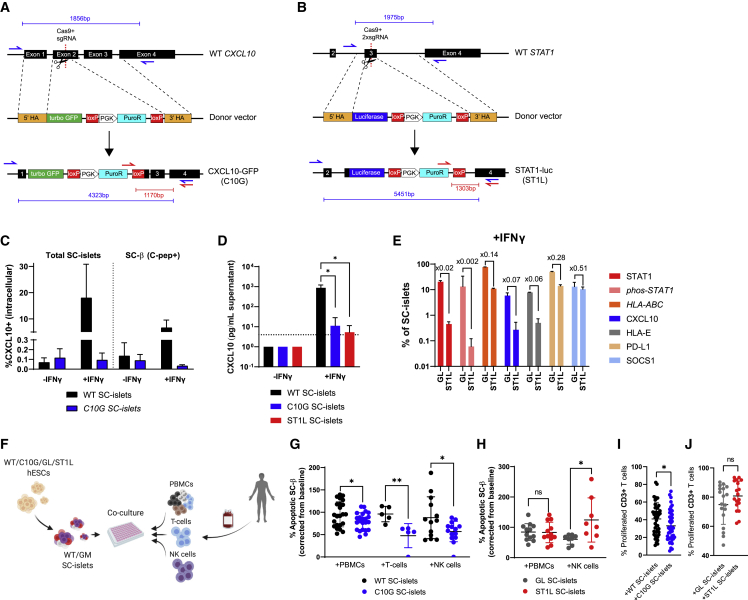
(A and B) Scheme of targeting the (A) CXCL10 or (B) STAT1 locus in hESCs using CRISPR. Red and blue arrows are PCR primers for genotyping as shown in Figure S4A.
(C) Flow cytometry of intracellular CXCL10 protein in WT/C10G SC-islets and SC-β (C-peptide+) ± rhIFNγ for 48 h n = 3–5.
(D) CXCL10 ELISA of supernatants from ±rhIFNγ-treated WT/C10G/ST1L SC-islets. Dashed line is the lower detection limit, while any data below it is extrapolated.
(E) Flow cytometry for protein expression in rhIFNγ-treated GAPDH-luciferase (GL) or ST1L SC-islets. n = 3–4.
(F–J) Gene-modified (GM; C10G/ST1L) and control (WT/GL) lines were differentiated into SC-islets, and co-cultured with human PBMCs or purified T cells/NK cells. Apoptosis was calculated by fraction from baseline (%TUNEL without PBMCs). (G) Apoptotic WT or C10G SC-β cells (n = 4 for ×6 PBMC donors, n = 2–3 ×2 T cell donors, n = 4 × 4 NK cell donors).
(H) Apoptotic GL or ST1L SC-β cells (n = 4 for ×2 PBMC or NK cell). (I and J) Proliferated CD3 T cell following co-culture with indicated GM SC-islets (I) n = 9 for ×5 donors and (J) n = 9 for ×2 donors). Error bars are mean ± SD. ns, not significant; ∗p < 0.05; ∗∗p < 0.01, unpaired two-tailed t-test.