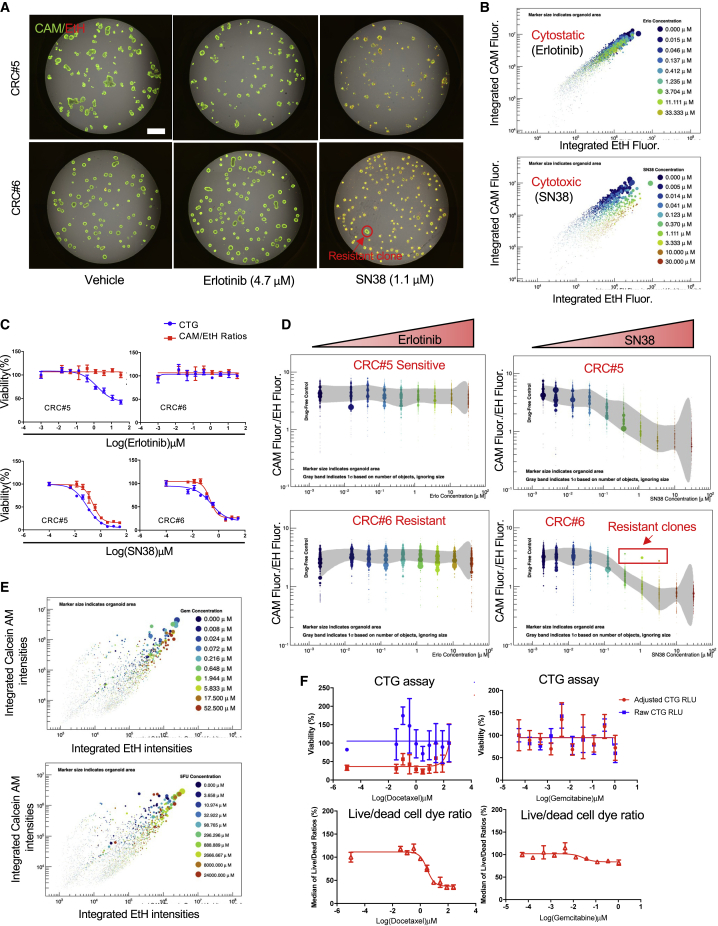
Figure 7.
An orthogonal AI-based analysis approach to differentiate the cytostatic/cytotoxic drug effects and capture heterogeneous drug response at individual-tumorsphere/organoid resolution
(A) Representative images of two CRC MOS treated with vehicle, erlotinib, or SN38, co-stained with live cell dye (CAM) and dead cell dye (EtH). The red circle highlights a resistant clone discovered in CRC#6 treated with SN38 (scale bar: 1,000 μm).
(B) Scatterplots show the differential drug responsive patterns of CRC#5 treated with of erlotinib (cytostatic) or SN38 (cytotoxic) respectively at the individual organoid resolution. The size of each dot reflects the relative surface area of the individual segmented object.
(C) The drug response curves of two CRC MOS models treated with erlotinib or SN38. Blue curves were plotted based on CTG assay and red curves were plotted based on the median ratios of CAM/EtH dye integrated intensities.
(D) Scatterplots show the dose-dependent changes of the ratios of CAM/EtH dye integrated intensities of CRC#6 and CRC#5 treated with erlotinib or SN38, respectively. The red rectangle highlights the drug-resistant clones identified in the CRC#6 treated with SN38. The x axis indicates the range of drug concentrations. The size of each dot reflects the relative surface area of the individual segmented object. Gray band indicates 1σ based on number of objects, ignoring size.
(E) Scatterplots show how the individual tumorspheres responded to 3 days of gemcitabine or 5-FU treatment in a primary lung tumor-derived MOS.
(F) The comparisons of drug response curves measured by CTG assay (top panel) versus median of live/dead cell dye ratios (bottom panel) in a sarcoma primary tissue-derived MOS.