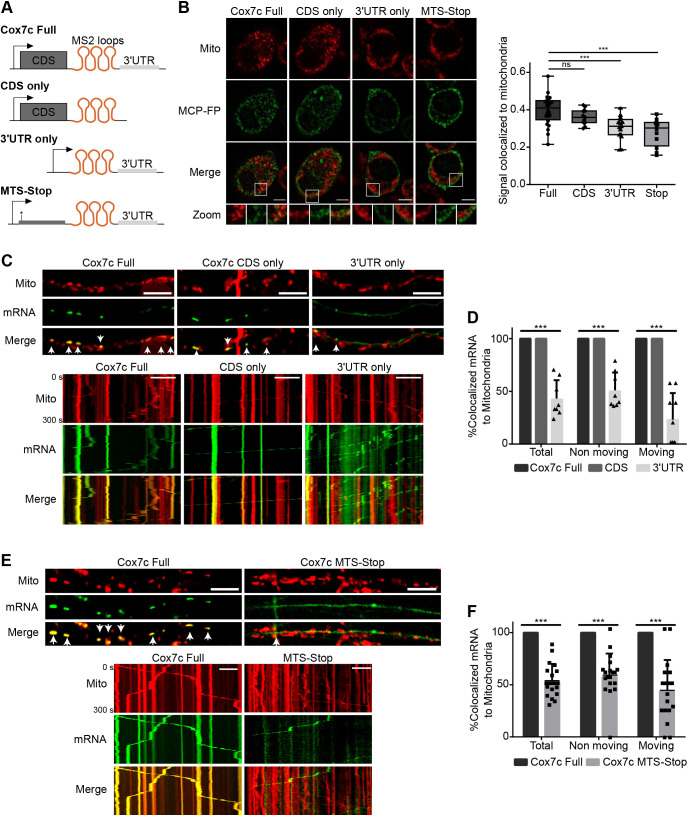
Fig. 3.
CDS determinants are important for Cox7c axonal mitochondrial association and cotransport. (A) Schematic representation of MS2-loops Cox7c Full, Cox7c CDS only, Cox7c 3′ UTR only or Cox7c MTS-Stop (a point mutation generating a stop codon closely downstream of the initiation codon) constructs. (B) Left, representative images of N2a cells transfected with the MS2 constructs with MS2 binding protein fused to YFP (MCP-FP; green). Scale bars: 5 μm. Right: quantification of the mRNA signal overlapping with mitochondria. Each data point represents a frame. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum and maximum values; n=42, 17, 25, 18 cells from 24, 11, 15, 12 frames for Cox7c Full, CDS only, 3′ UTR and MTS-Stop, respectively. n.s. (non-significant) P>0.05, ***P<0.001 (Mann–Whitney test). (C) Representative images (top) and kymographs (bottom) of primary motor neurons infected with viral vectors expressing Cox7c Full, Cox7c CDS only or Cox7c 3′ UTR only and MCP–GFP (green). Axons show axonal colocalization with mitochondria (red) in the Full and CDS only but not in the 3′ UTR only construct. Arrowheads indicate colocalization of mitochondria and mRNA signals. Scale bars: 10 μm. (D) Quantification of axonal cotransport of mRNA and mitochondria, separated according to moving and static mRNA. Error bars are s.d.; n=8, 12, 8 axons from three biological independent cultures. ***P<0.001 (two-way ANOVA with Holm–Sidak correction). (E) Representative images and kymographs of primary motor neurons infected with viral vectors expressing Full Cox7c or Cox7c ‘STOP’ and MCP–GFP (green). Axons show axonal colocalization with mitochondria (red) in the Full Cox7c but not in the Cox7c ‘STOP’ construct. Arrowheads indicate colocalization of mitochondria and mRNA signals. Scale bars: 10 μm. (F) Quantification of axonal cotransport of mRNA and mitochondria, separated according to moving and static mRNA. Error bars are s.d.; n=9 and 19 axons from three biological independent cultures. ***P<0.001 (two-way ANOVA with Holm–Sidak correction).