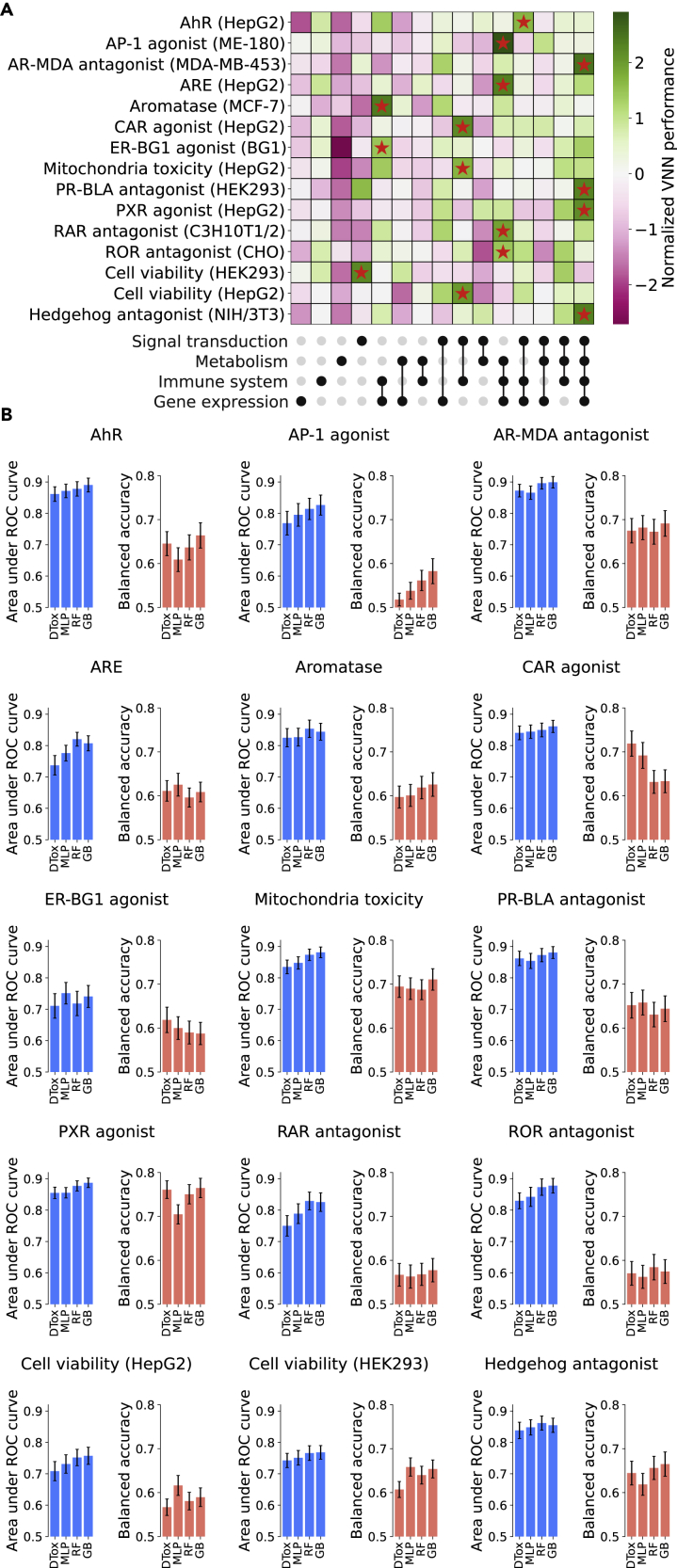
Figure 2.
Prediction of compound response to 15 toxicity assays
(A) Heatmap showing the training performance of VNN built under different combinations of root biological processes (shown as upset plot at the bottom). To facilitate comparison, the model performance is normalized within each assay using Z-transform. The optimal combination for each assay is highlighted with a red star. The name of each assay is annotated on the left, with the name of the assay cell line included in parentheses.
(B) Bar plot showing the validation performance in all 15 Tox21 datasets. The performance of DTox is compared against three other models: a multi-layer perceptron with the same number of hidden layers and neurons as DTox (MLP), random forest (RF), and gradient boosting (GB). Performance is measured by two metrics: area under ROC curve and balanced accuracy, with error bar showing the 95% confidence interval.
AhR, aryl hydrocarbon receptor; AP-1, activator protein-1; ARE, antioxidant response element; AR-MDA, androgen receptor in MDA-kb2 AR-luc cell line; CAR, constitutive androstane receptor; ER-BG1, estrogen receptor in BG1 cell line, PR-BLA, progesterone receptor in PR-UAS-bla HEK293T cell line; PXR, pregnane X receptor; RAR, retinoid acid receptor; ROR, retinoid-related orphan receptor.