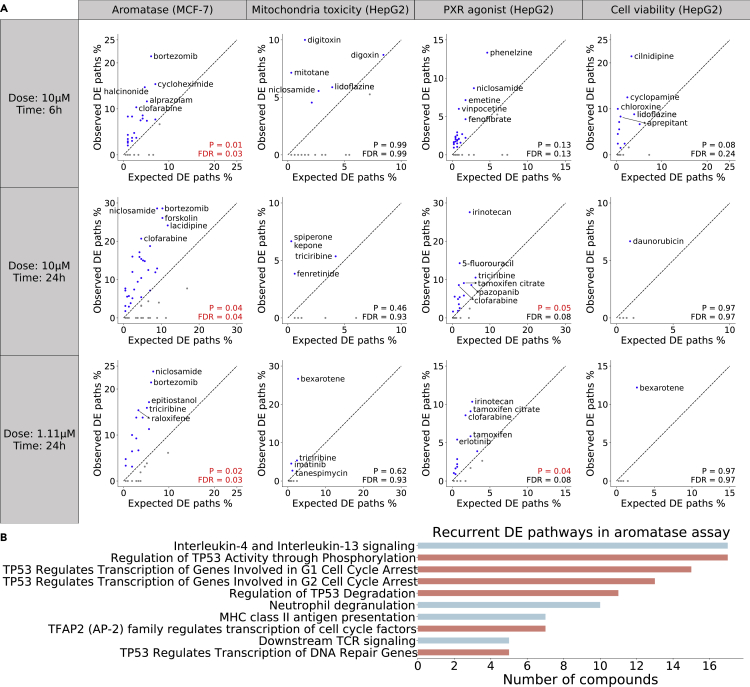
Figure 4.
Validation of identified VNN paths by differential expression
(A) A VNN path is considered to be differentially expressed (DE) if all pathways along the path are enriched for DE genes from the matched LINCS experiment. The validation analysis is performed for four assays (columns) in three different dose-time groups (rows), with each scatterplot comparing the observed versus expected proportion of DE paths for a single group. The observed proportion is computed with VNN paths identified for each compound, while the expected proportion is computed with all possible VNN paths. A Wilcoxon signed-rank test is employed to examine whether the average observed proportion of each group is significantly higher than the average expected proportion (p value and FDR shown at the bottom right). The diagonal is shown as black dashed line, with compounds in the upper triangle (observed > expected) shown in blue and compounds in the lower triangle (observed < expected) shown in gray. Compounds with the top five observed proportions in each group are annotated with their names.
(B) Bar plot showing the DE VNN paths that are recurrently identified for at least five aromatase inhibitors. Each VNN path is named after its lowest-level pathway. Paths that contain the “transcriptional regulation by TP53” pathway are highlighted in salmon, while the remaining paths are colored in cyan.