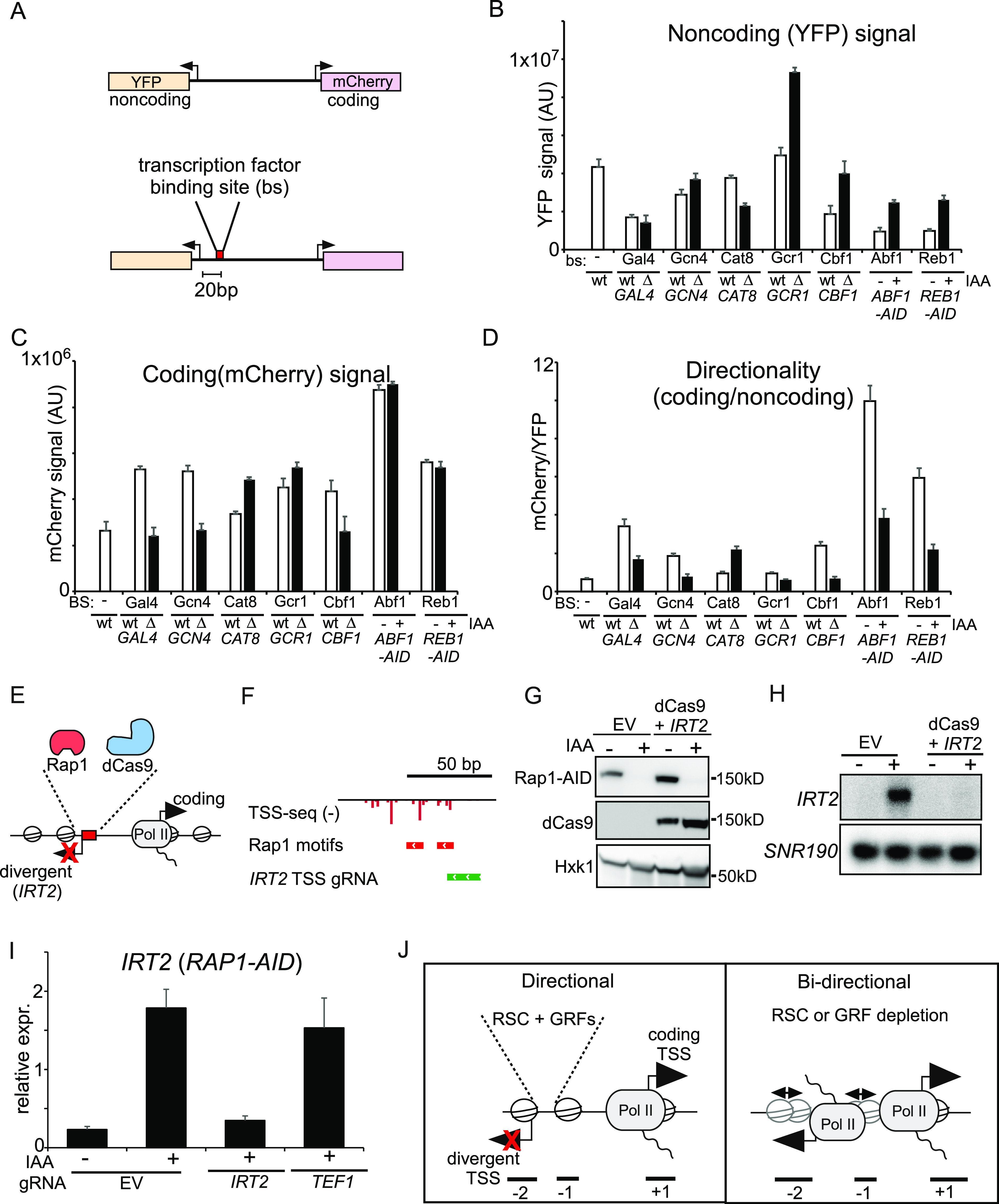
Figure 6. Targeting general regulatory factors or dCas9 to divergent core promoters is sufficient to repress divergent noncoding transcription.
(A) Schematic overview of the reporter construct. The transcription factor binding sites were cloned 20 nucleotides upstream of the YFP (SUT129) transcription start site (TSS). (B) YFP, noncoding, signal for constructs harbouring binding sites for Gal4, Gcn4, Cat8, Gcr1, Cbf1, Abf1, and Reb1 cloned into the WT PPT1 promoter (FW6407). The YFP activity was determined in WT control cells and in gene deletion strains of matching transcription factor binding site reporter constructs (FW6404, FW6306, FW6401, FW6300, FW6402, FW6302, FW6403, FW6424, FW6405, and FW6315). For Abf1 and Reb1 reporter constructs, Abf1 and Reb1 were depleted using the auxin-inducible degron (ABF1-AID and REB1-AID) (FW6415 and FW6411). Cells were treated for 2 h with IAA or DMSO. Displayed is the mean signal of at least 50 cells. The error bars represent 95% confidence intervals. (C) Same analysis as B except that coding direction is shown. (B, D) Same analysis as (B) except that directionality was calculated by taking the ratio of coding over noncoding. (E) Scheme depicting the targeting of dCas9 to repress the divergent transcript IRT2. (F) Design of gRNAs targeting the IRT2 TSS. Displayed are TSS-seq data from after Rap1 depletion (Wu et al, 2018). (G) Western blot of Rap1 and dCas9 detected with anti-V5 and anti-FLAG antibodies, respectively. Cells harbouring RAP1-AID were treated with IAA to deplete Rap1 and induce IRT2 expression in either cells with empty vector (EV) or cells harbouring a dCas9 construct and a construct expressing the gRNA targeted to the Rap1-binding site (FW8477, FW8531). Hxk1 was used as a loading control, detected using anti-Hxk1 antibodies. (E, H) IRT2 expression detected by Northern blot for strains and treatments described in (E). As a loading control, the membrane was re-probed for SNR190. (D, I) IRT2 expression as detected in by qRT-PCR of as described in (D). A control gRNA was included in the analysis targeted to the TSS of TEF1 (FW8527). Displayed are the mean signals of n = 3 biological repeats and SEM. (J) Model for promoter directionality.
Source data are available for this figure.