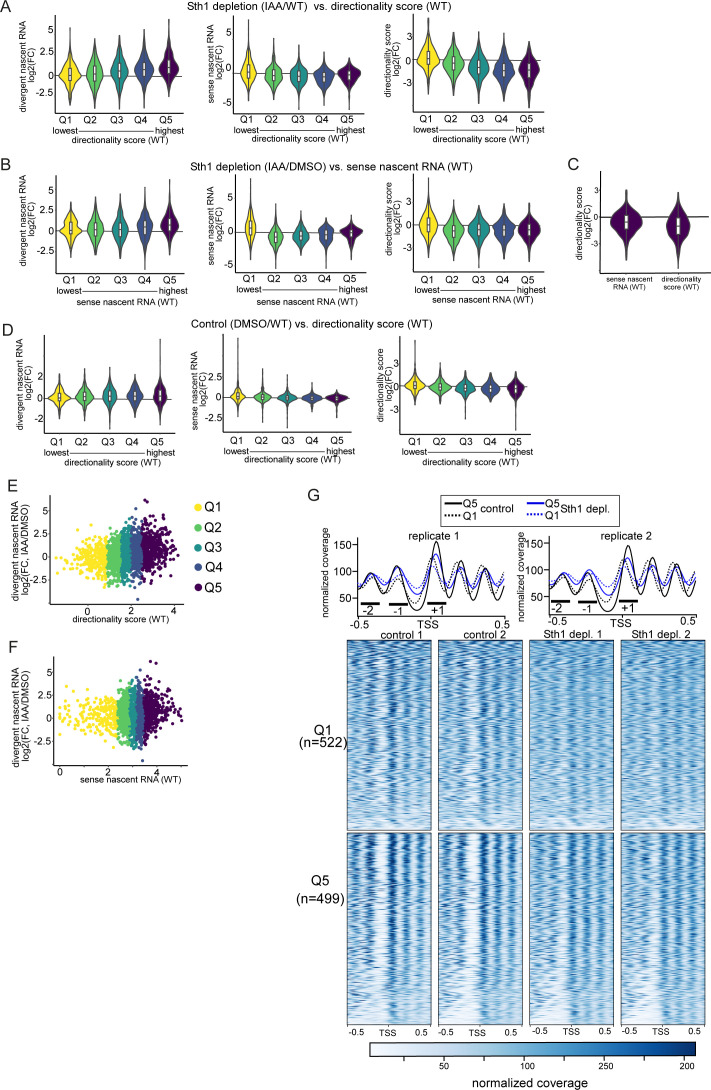
Figure S4. RSC acts at highly directional promoters.
(A) Violin plots displaying the distribution of changes in divergent transcription (left), sense transcription (centre), and directionality score (right) in Sth1-depleted versus WT cells (log2(FC) IAA/WT), in sets of tandem genes stratified according to their directionality in WT cells (x-axis, Q1 to Q5). Q1 and Q5, respectively, show the group of gene promoters with lowest and highest directionality score in WT cells. The range for directionality scores for each group are the following: Q1(−3.11–0.85), Q2(0.86–1.49), Q3(1.50–1.97), Q4(1.98–2.48), Q5(2.49–4.20). (B) Violin plots displaying the distribution of changes in divergent transcription (left), sense transcription (centre), and directionality score (right) in Sth1-depleted versus control cells (log2(FC) IAA/DMSO), in sets of tandem genes stratified according to their sense signal in WT cells (x-axis, Q1 to Q5). Q1 and Q5, respectively, show the group of gene promoters with lowest and highest directionality score in WT cells. The range for directionality scores for each group are the following: Q1(0.99–239), Q2(240–746), Q3(747–1,440), Q4(1,441–2,848), Q5(2,849–105.685). Average values between replicates (n = 3) are represented. (C) Violin plots showing the distribution of changes in divergent transcription (log2(FC) IAA/DMSO) in either the set of gene promoters with the highest transcription (Q5 sense WT, as in Fig S4B) or the set of gene promoters with the highest directionality scores (Q5 directionality WT, as in Fig 4A). (D) Similar as (A), but displayed is the comparison of control versus WT (DMSO/WT) sorted on directionality score. (E) Same data as in Fig 4A (left panel), but the data are represented by a scatter plot. (F) Same data as in Fig S4B (left panel), but the data are represented by a scatter plot. (G) Metagene and heatmap analysis of MNase-seq data obtained from Klein-Brill et al (2019) for genes belonging to directionality Q1 and Q5. Plots are centred on coding gene transcription start site as in Fig 4C and D, and replicates (n = 2) are shown separately. Marked are the regions representing the +1, −1, and −2 nucleosome positions.