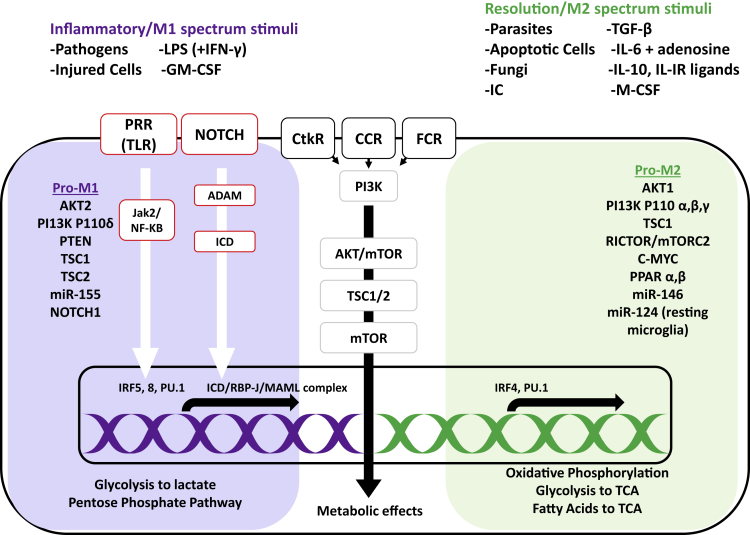
Figure 6.
Key stimuli and molecular pathways in macrophages with the inflammatory (M1) versus healing phenotype (M2)
The M1 phenotype is activated by signals from pathogens, injured cells, and in vitro stimuli, and the M2 phenotype is activated by parasites, fungi, apoptotic cells, immune complexes, and other cytokine/growth factor stimuli. Signals that are sensed via pathogen-recognition receptors (PRRs), such as Toll-like receptors (TLRs), result in Janus-activated kinase (Jak) 2 and nuclear factor κB (NF-κB) activation. Signals received via Notch receptors, cytokine receptor (CtkR), chemokine receptor (CCR), and Fc receptor (FcR) stimulation also define gene expression and downstream metabolic reprogramming. IRF5 and IRF8 induce inflammatory M1 gene expression, whereas IRF 4 promotes M2 phenotype genes that, in turn, promote changes in nutrient uptake and metabolic pathways, depending on which part of the gene is expressed. LPS, lipopolysaccharide; GM-CSF, granulocyte monocyte colony-stimulating factor; IC, immune complex; M-CSF, monocyte colony-stimulating factor; PI3K, phosphatidylinositol 3-kinase; AKT, serine threonine kinase; mTOR, mammalian target of rapamycin; PTEN, phosphatase and tensin homolog; TSC, tuberous sclerosis complex; PPAR, peroxisome proliferator-activated receptor; ADAM, a disintegrin and metalloproteinase; RBP-J, recombination signal binding protein for immunoglobulin κJ region; MAML, mastermind-like; Rictor, rapamycin-insensitive companion of mTOR; TCA, tricarboxylic acid/Krebs cycle. Adapted from Amici et al.113