Figure 7.
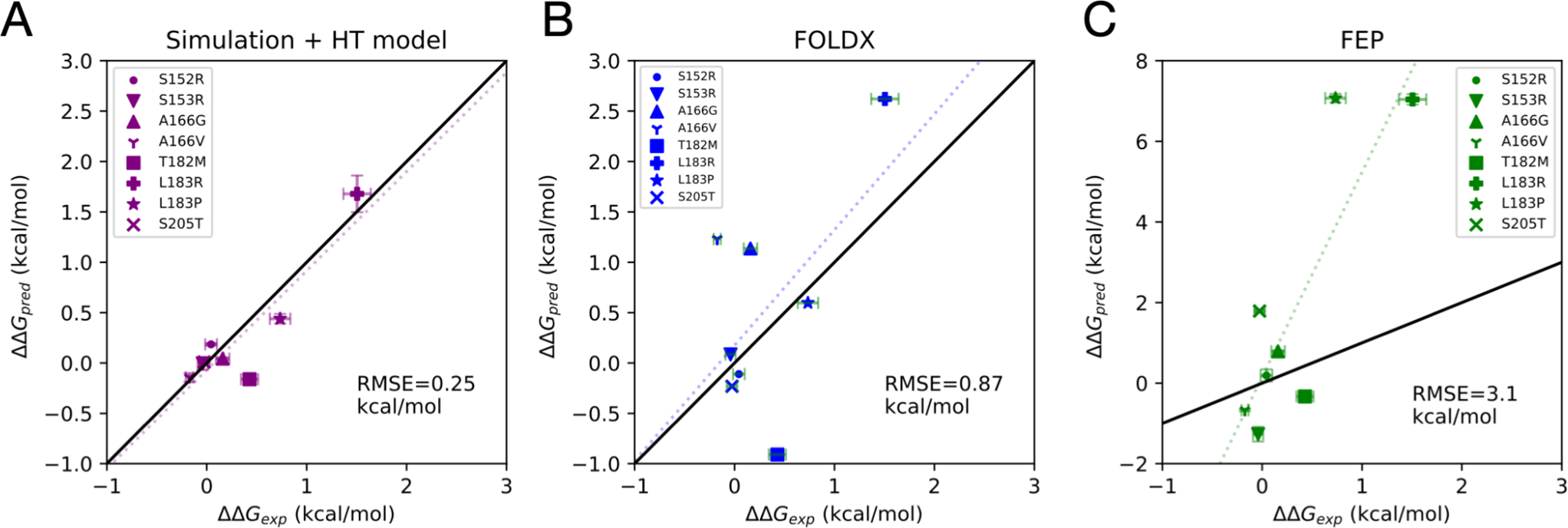
Comparison of estimated experimental folding free energy changes upon mutation ΔΔGexp, and predicted estimates ΔΔGpred from three different computational methods: (A) a combined molecular simulation and hydrophobic transfer (HT) model, (B) predictions from the FoldX algorithm, and (C) alchemical FEP estimation. In each plot, the dotted line is the least-squares linear fit, and the root mean squared error (RMSE).
