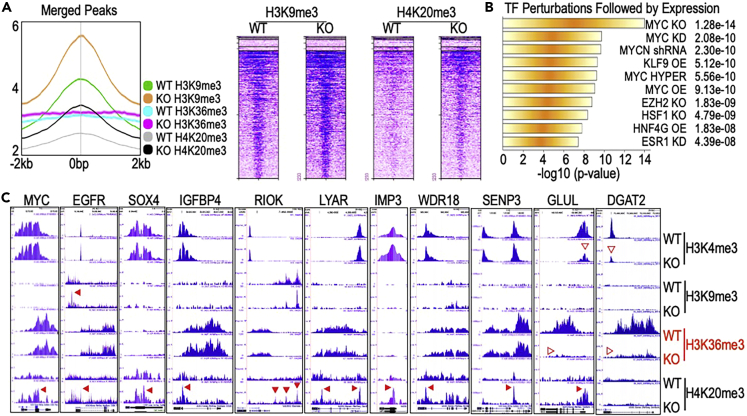
Figure 6.
Enhanced repressive histone trimethylation and down-regulation of MYC signaling in the mdig KO cells
(A) Average plots of the merged peaks of ChIP-seq for H3K9me3, H3K36me3 and H4K20me3 in the regions from -2 kb to 2 kb of gene loci in WT and mdig KO cells. Right panel shows heatmaps of the merged peak distribution of H3K9me3 and H4K20me3 between WT and mdig KO cells.
(B) Transcription factor pathway analysis using Enrichr TF Perturbations Followed by Expression for the 980 downregulated proteins in the mdig KO MDA-MB-231 cells.
(C) Histone trimethylation profiles of the selected MYC pathway genes in both WT and mdig KO cells. Block arrowheads depict the enhanced enrichment of the repressive histone trimethylation markers on the indicated genes in KO cells. Empty arrowheads indicate diminished active histone trimethylation markers in the indicated gene in WT and KO cells.